Chapter 13 MI50: Multi-path exploration results
Here we present the results for the best performances and activation gene coverage generated by each selection scheme replicate on the multi-path exploration diagnostic with configurations presented below. For our the configuration of these experiments, we execute migrations every 50 generations and there are 4 islands in a ring topology. Best performance found refers to the largest average trait score found in a given population. Note that activation gene coverage values are gathered at the population-level. Activation gene coverage refers to the count of unique activation genes in a given population; this gives us a range of integers between 0 and 100.
13.2 Truncation selection
Here we analyze how the different population structures affect truncation selection (size 8) on the contradictory objectives diagnostic.
13.2.1 Performance
13.2.1.1 Performance over time
lines = filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TRUNCATION') %>%
group_by(Structure, Generations) %>%
dplyr::summarise(
min = min(pop_fit_max) / DIMENSIONALITY,
mean = mean(pop_fit_max) / DIMENSIONALITY,
max = max(pop_fit_max) / DIMENSIONALITY
)
ggplot(lines, aes(x=Generations, y=mean, group = Structure, fill = Structure, color = Structure, shape = Structure)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, Generations %% 2000 == 0), size = 2.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Performance over time") +
p_theme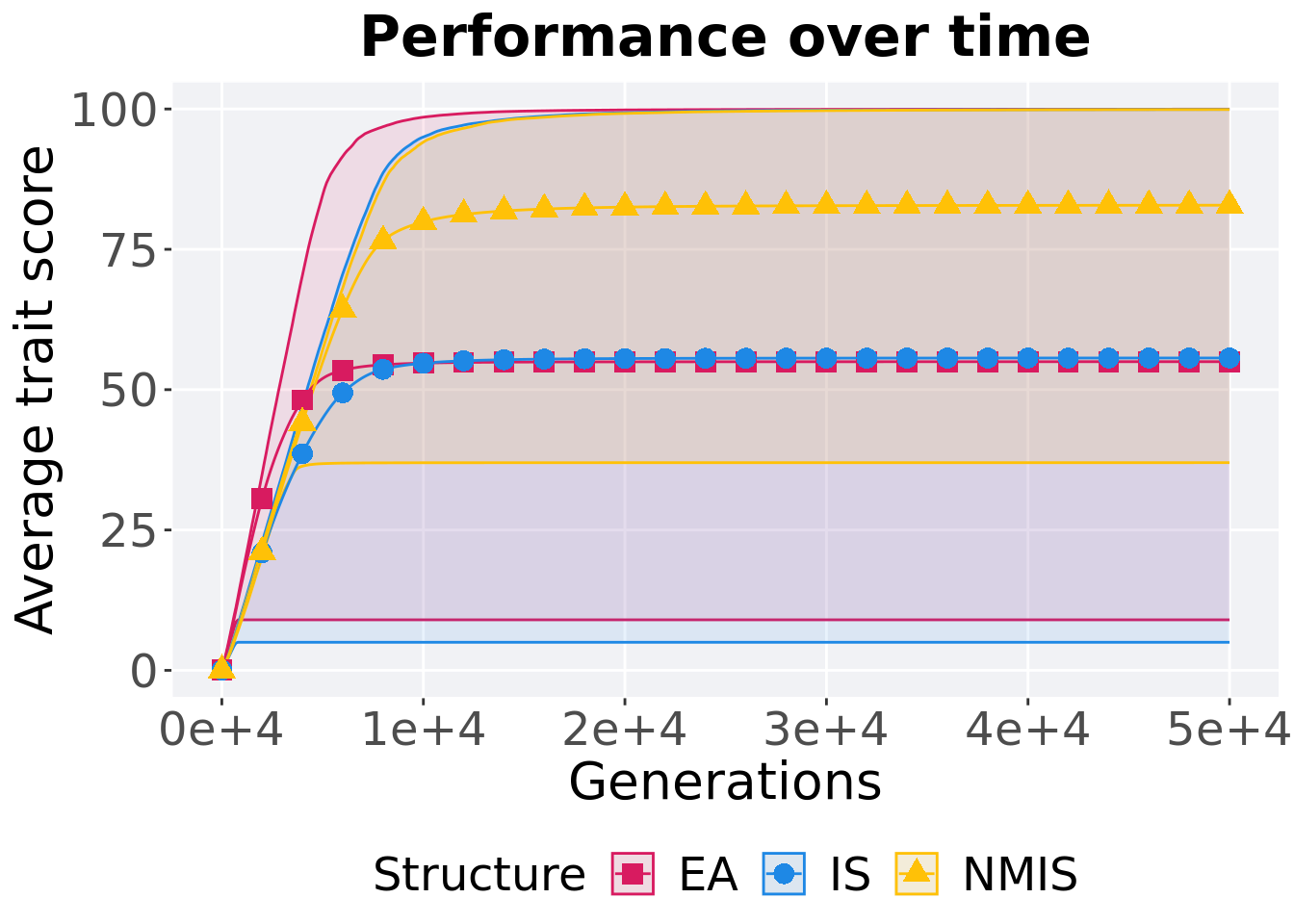
13.2.1.2 Best performance
First generation a satisfactory solution is found throughout the 50,000 generations.
filter(mi50_best, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TRUNCATION' & VAR == 'pop_fit_max') %>%
ggplot(., aes(x = Structure, y = VAL / DIMENSIONALITY, color = Structure, fill = Structure, shape = Structure)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_discrete(
name="Structure"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette, ) +
scale_fill_manual(values = cb_palette) +
ggtitle('Best performance')+
p_theme + coord_flip()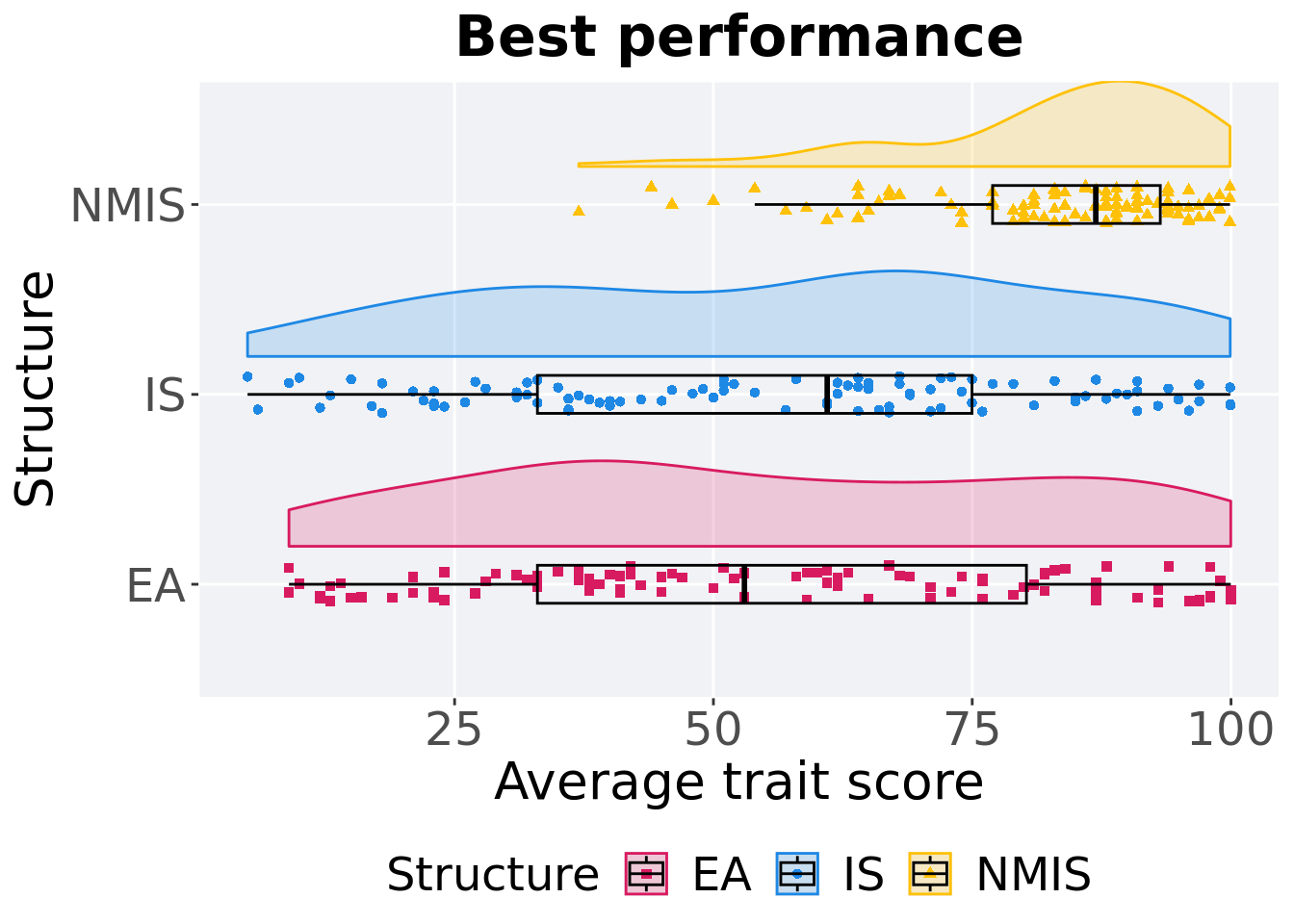
13.2.1.2.1 Stats
Summary statistics for the first generation a satisfactory solution is found.
performance = filter(mi50_best, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TRUNCATION' & VAR == 'pop_fit_max')
performance %>%
group_by(Structure) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(VAL)),
min = min(VAL, na.rm = TRUE) / DIMENSIONALITY,
median = median(VAL, na.rm = TRUE) / DIMENSIONALITY,
mean = mean(VAL, na.rm = TRUE) / DIMENSIONALITY,
max = max(VAL, na.rm = TRUE) / DIMENSIONALITY,
IQR = IQR(VAL, na.rm = TRUE) / DIMENSIONALITY
)## # A tibble: 3 x 8
## Structure count na_cnt min median mean max IQR
## <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 EA 100 0 9.00 53.0 55.0 100. 47.2
## 2 IS 100 0 5 61.0 55.6 99.9 42.0
## 3 NMIS 100 0 37.0 86.9 82.9 99.9 16.2Kruskal–Wallis test provides evidence of difference among selection schemes.
##
## Kruskal-Wallis rank sum test
##
## data: VAL by Structure
## Kruskal-Wallis chi-squared = 67.87, df = 2, p-value = 1.829e-15Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction.
pairwise.wilcox.test(x = performance$VAL, g = performance$Structure, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'g')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: performance$VAL and performance$Structure
##
## EA IS
## IS 1 -
## NMIS 3.0e-12 7.9e-13
##
## P value adjustment method: bonferroni13.2.1.3 Final performance
First generation a satisfactory solution is found throughout the 50,000 generations.
filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TRUNCATION' & Generations == 50000) %>%
ggplot(., aes(x = Structure, y = pop_fit_max / DIMENSIONALITY, color = Structure, fill = Structure, shape = Structure)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_discrete(
name="Structure"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette, ) +
scale_fill_manual(values = cb_palette) +
ggtitle('Final performance')+
p_theme + coord_flip()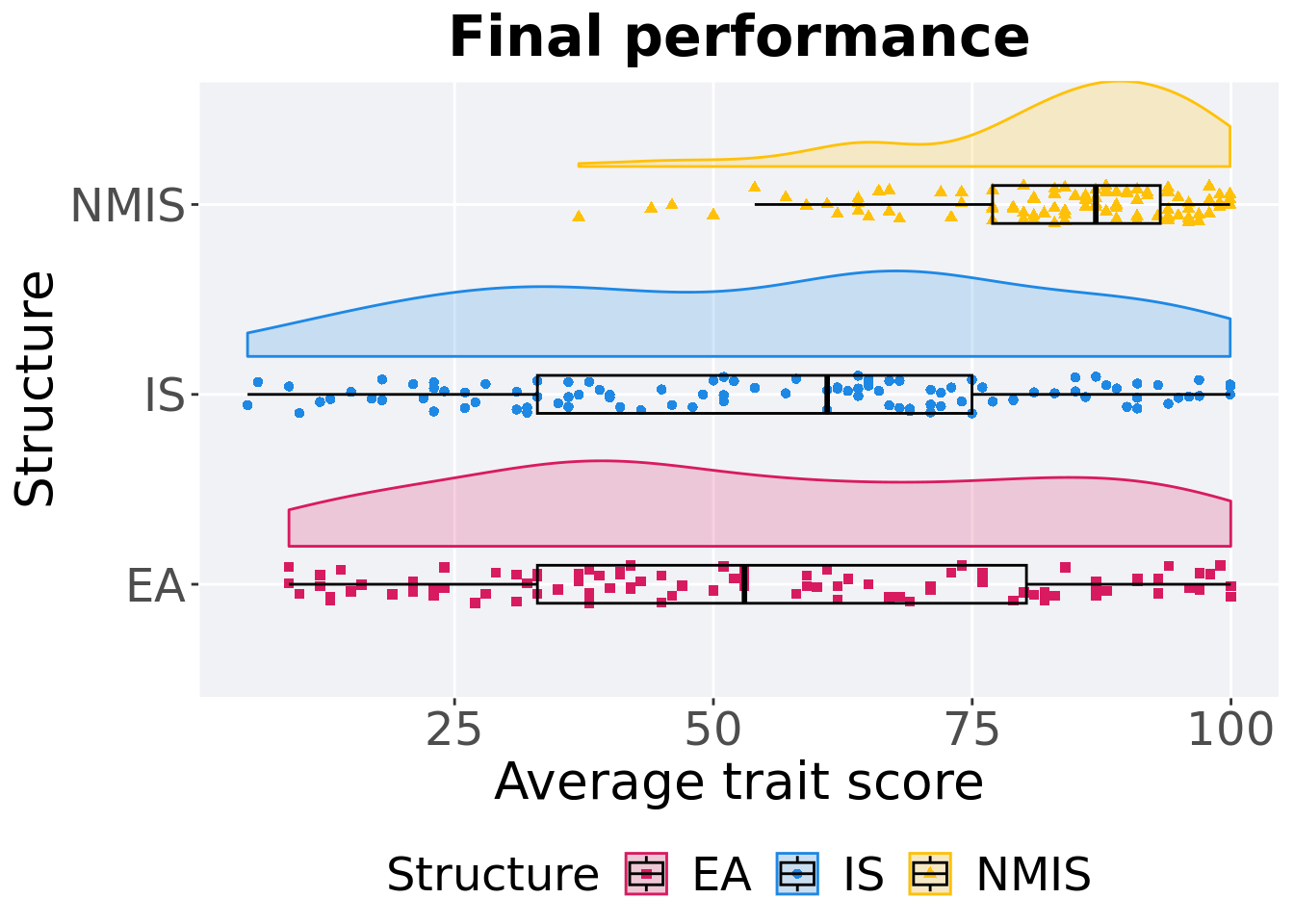
13.2.1.3.1 Stats
Summary statistics for the first generation a satisfactory solution is found.
performance = filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TRUNCATION' & Generations == 50000)
performance %>%
group_by(Structure) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_fit_max)),
min = min(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
median = median(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
mean = mean(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
max = max(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
IQR = IQR(pop_fit_max / DIMENSIONALITY, na.rm = TRUE)
)## # A tibble: 3 x 8
## Structure count na_cnt min median mean max IQR
## <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 EA 100 0 9.00 53.0 55.0 100. 47.2
## 2 IS 100 0 5 61.0 55.6 99.9 42.0
## 3 NMIS 100 0 37.0 86.9 82.9 99.9 16.2Kruskal–Wallis test provides evidence of difference among selection schemes.
##
## Kruskal-Wallis rank sum test
##
## data: pop_fit_max by Structure
## Kruskal-Wallis chi-squared = 67.87, df = 2, p-value = 1.829e-15Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction.
pairwise.wilcox.test(x = performance$pop_fit_max, g = performance$Structure, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'g')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: performance$pop_fit_max and performance$Structure
##
## EA IS
## IS 1 -
## NMIS 3.0e-12 7.9e-13
##
## P value adjustment method: bonferroni13.2.2 Generation satisfactory solution found
First generation a satisfactory solution is found throughout the 50,000 generations.
filter(mi50_ssf, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TRUNCATION'& Generations <= GENERATIONS) %>%
ggplot(., aes(x = Structure, y = Generations, color = Structure, fill = Structure, shape = Structure)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_shape_manual(values=SHAPE)+
scale_y_continuous(
name="Generations"
) +
scale_x_discrete(
name="Structure"
) +
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
p_theme + coord_flip()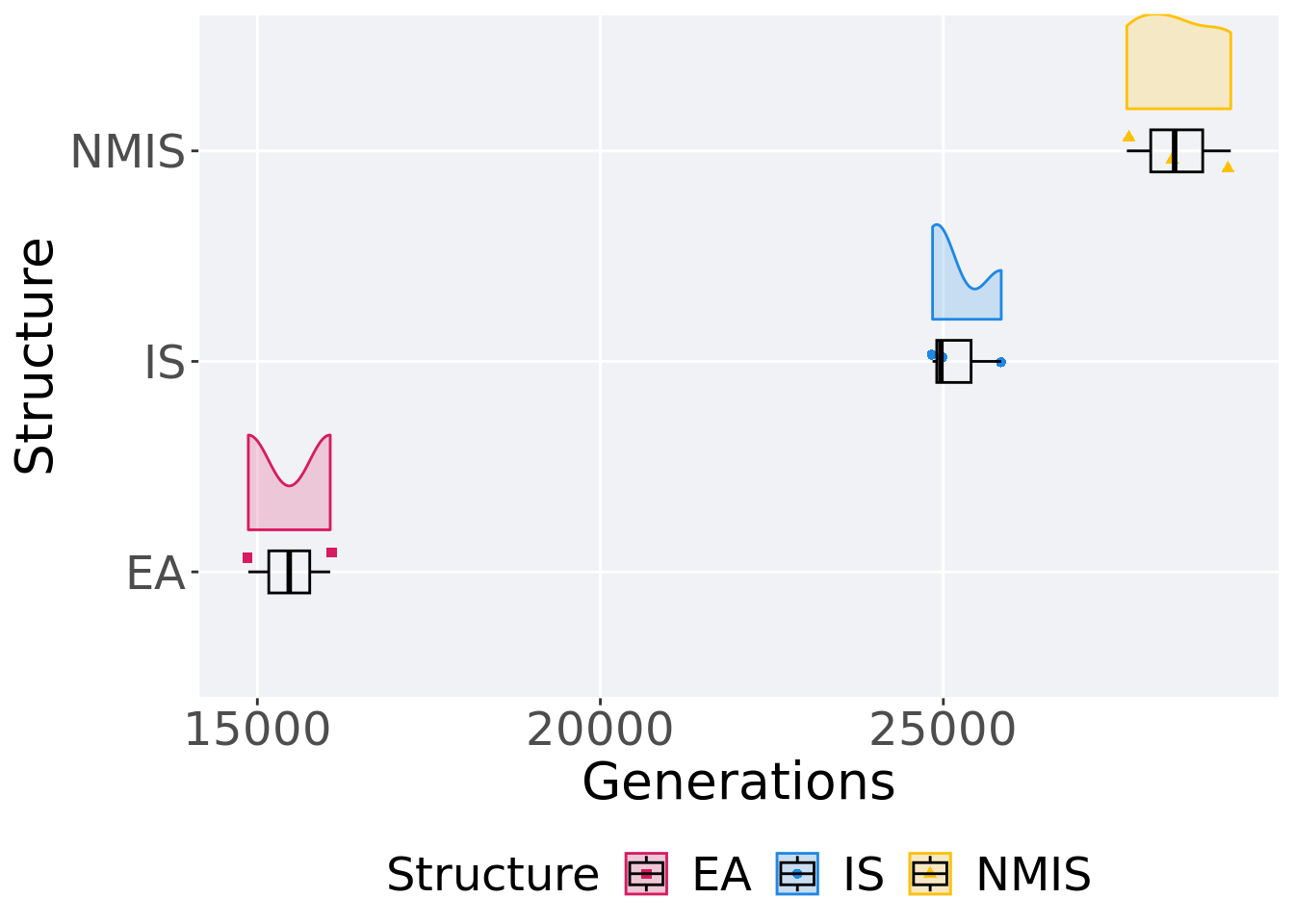
13.2.2.1 Stats
Summary statistics for the first generation a satisfactory solution is found.
ssf = filter(mi50_ssf, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TRUNCATION' & Generations < 60000)
ssf %>%
group_by(Structure) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(Generations)),
min = min(Generations, na.rm = TRUE),
median = median(Generations, na.rm = TRUE),
mean = mean(Generations, na.rm = TRUE),
max = max(Generations, na.rm = TRUE),
IQR = IQR(Generations, na.rm = TRUE)
)## # A tibble: 3 x 8
## Structure count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 EA 2 0 14868 15465 15465 16062 597
## 2 IS 3 0 24843 24965 25217. 25844 500.
## 3 NMIS 3 0 27675 28372 28412. 29190 758.Kruskal–Wallis test provides evidence of no difference among selection schemes.
##
## Kruskal-Wallis rank sum test
##
## data: Generations by Structure
## Kruskal-Wallis chi-squared = 6.25, df = 2, p-value = 0.0439413.2.3 Activation gene coverage
Activation gene coverage analysis.
13.2.3.1 Coverage over time
Activation gene coverage over time.
# data for lines and shading on plots
lines = filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TRUNCATION') %>%
group_by(Structure, Generations) %>%
dplyr::summarise(
min = min(pop_act_cov),
mean = mean(pop_act_cov),
max = max(pop_act_cov)
)## `summarise()` has grouped output by 'Structure'. You can override using the
## `.groups` argument.ggplot(lines, aes(x=Generations, y=mean, group = Structure, fill = Structure, color = Structure, shape = Structure)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, Generations %% 2000 == 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Activation gene coverage over time')+
p_theme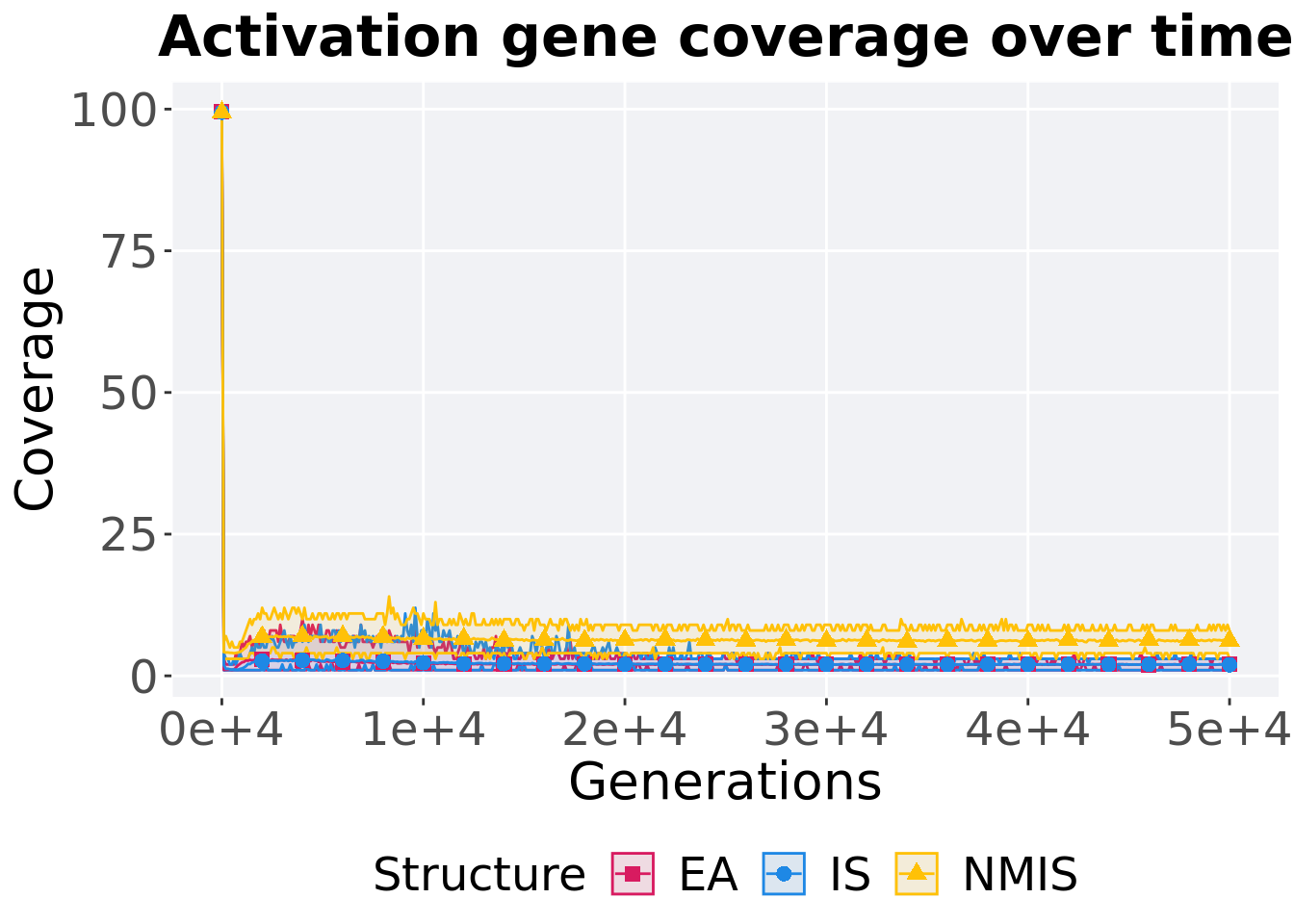
13.2.3.2 End of 50,000 generations
Activation gene coverage in the population at the end of 50,000 generations.
### end of run
filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TRUNCATION' & Generations == 50000) %>%
ggplot(., aes(x = Structure, y = pop_act_cov, color = Structure, fill = Structure, shape = Structure)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.3) +
geom_point(position = position_jitter(height = .05, width = .05), size = 1.5, alpha = 0.5) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_shape_manual(values=SHAPE)+
scale_y_continuous(
name="Coverage"
) +
scale_x_discrete(
name="Structure"
) +
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Final activation gene coverage')+
p_theme + coord_flip()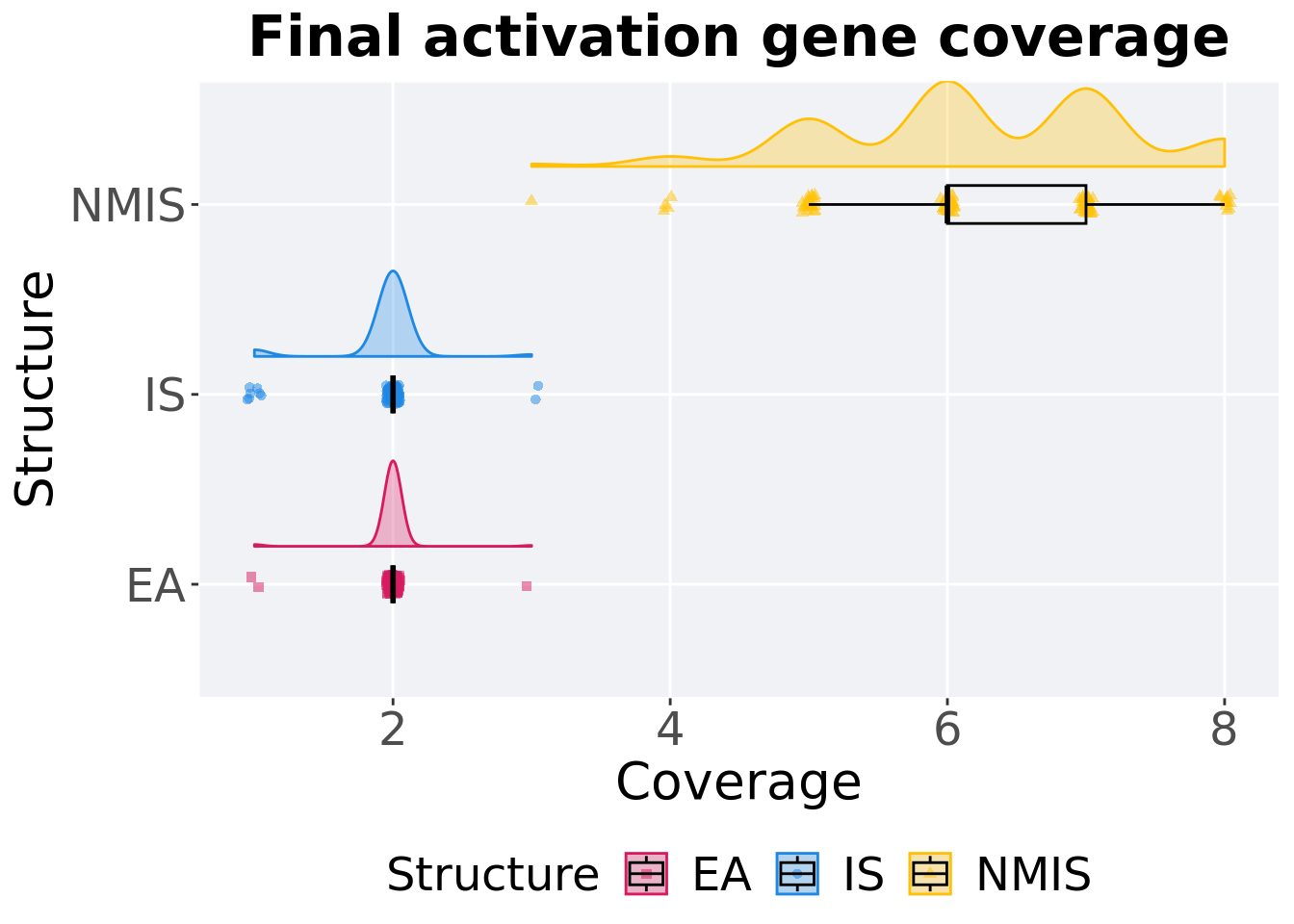
13.2.3.2.1 Stats
Summary statistics for activation gene coverage.
coverage = filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TRUNCATION' & Generations == 50000)
coverage %>%
group_by(Structure) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_act_cov)),
min = min(pop_act_cov, na.rm = TRUE),
median = median(pop_act_cov, na.rm = TRUE),
mean = mean(pop_act_cov, na.rm = TRUE),
max = max(pop_act_cov, na.rm = TRUE),
IQR = IQR(pop_act_cov, na.rm = TRUE)
)## # A tibble: 3 x 8
## Structure count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 EA 100 0 1 2 1.99 3 0
## 2 IS 100 0 1 2 1.95 3 0
## 3 NMIS 100 0 3 6 6.23 8 1Kruskal–Wallis test provides evidence of difference among activation gene coverage.
##
## Kruskal-Wallis rank sum test
##
## data: pop_act_cov by Structure
## Kruskal-Wallis chi-squared = 265.48, df = 2, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction on activation gene coverage.
pairwise.wilcox.test(x = coverage$pop_act_cov, g = coverage$Structure, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'g')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: coverage$pop_act_cov and coverage$Structure
##
## EA IS
## IS 1 -
## NMIS <2e-16 <2e-16
##
## P value adjustment method: bonferroni13.3 Tournament selection
Here we analyze how the different population structures affect tournament selection (size 8) on the contradictory objectives diagnostic.
13.3.1 Performance
13.3.1.1 Performance over time
lines = filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TOURNAMENT') %>%
group_by(Structure, Generations) %>%
dplyr::summarise(
min = min(pop_fit_max) / DIMENSIONALITY,
mean = mean(pop_fit_max) / DIMENSIONALITY,
max = max(pop_fit_max) / DIMENSIONALITY
)
ggplot(lines, aes(x=Generations, y=mean, group = Structure, fill = Structure, color = Structure, shape = Structure)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, Generations %% 2000 == 0), size = 2.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Performance over time") +
p_theme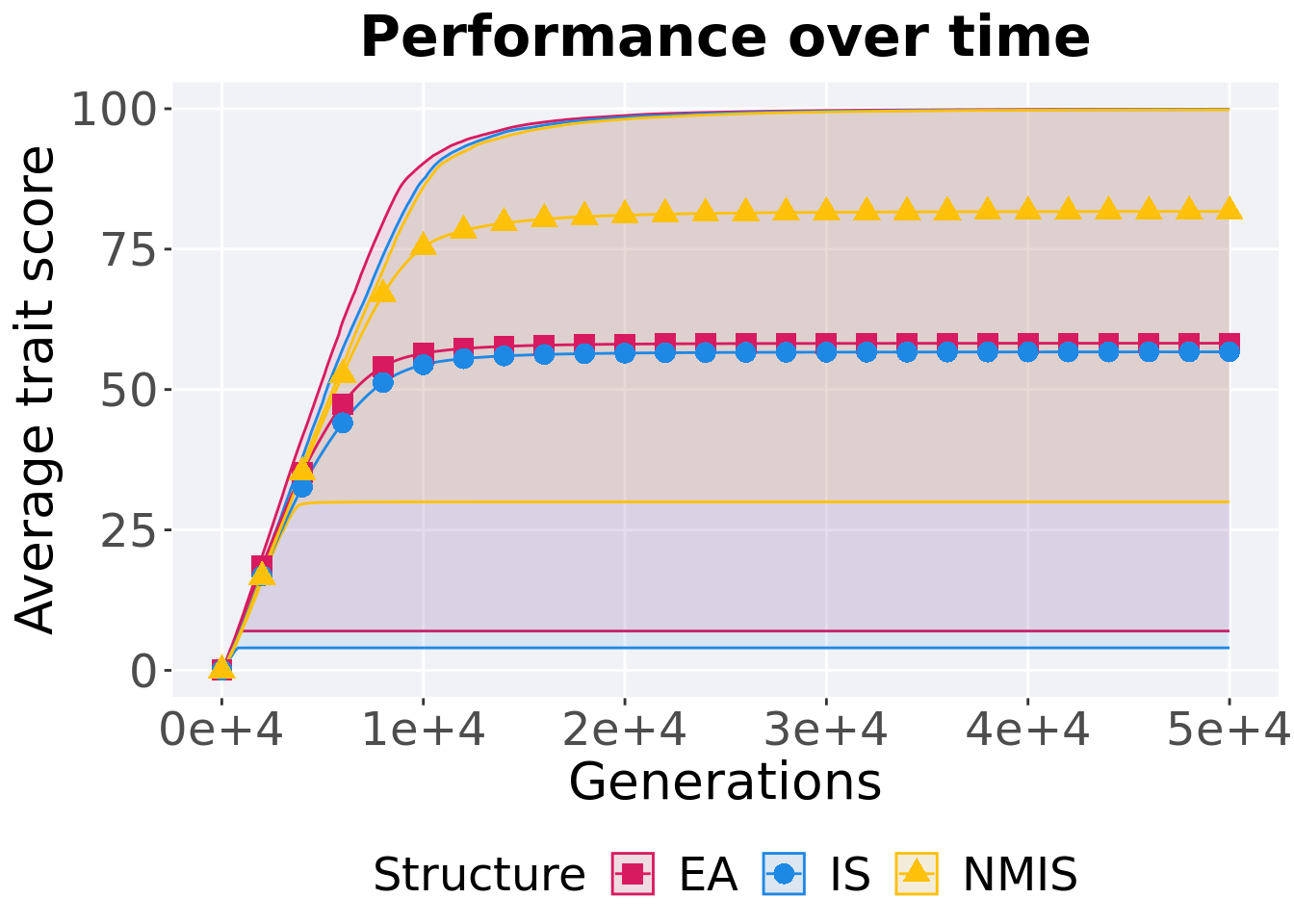
13.3.1.2 Best performance
First generation a satisfactory solution is found throughout the 50,000 generations.
filter(mi50_best, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TOURNAMENT' & VAR == 'pop_fit_max') %>%
ggplot(., aes(x = Structure, y = VAL / DIMENSIONALITY, color = Structure, fill = Structure, shape = Structure)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_discrete(
name="Structure"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette, ) +
scale_fill_manual(values = cb_palette) +
ggtitle('Best performance')+
p_theme + coord_flip()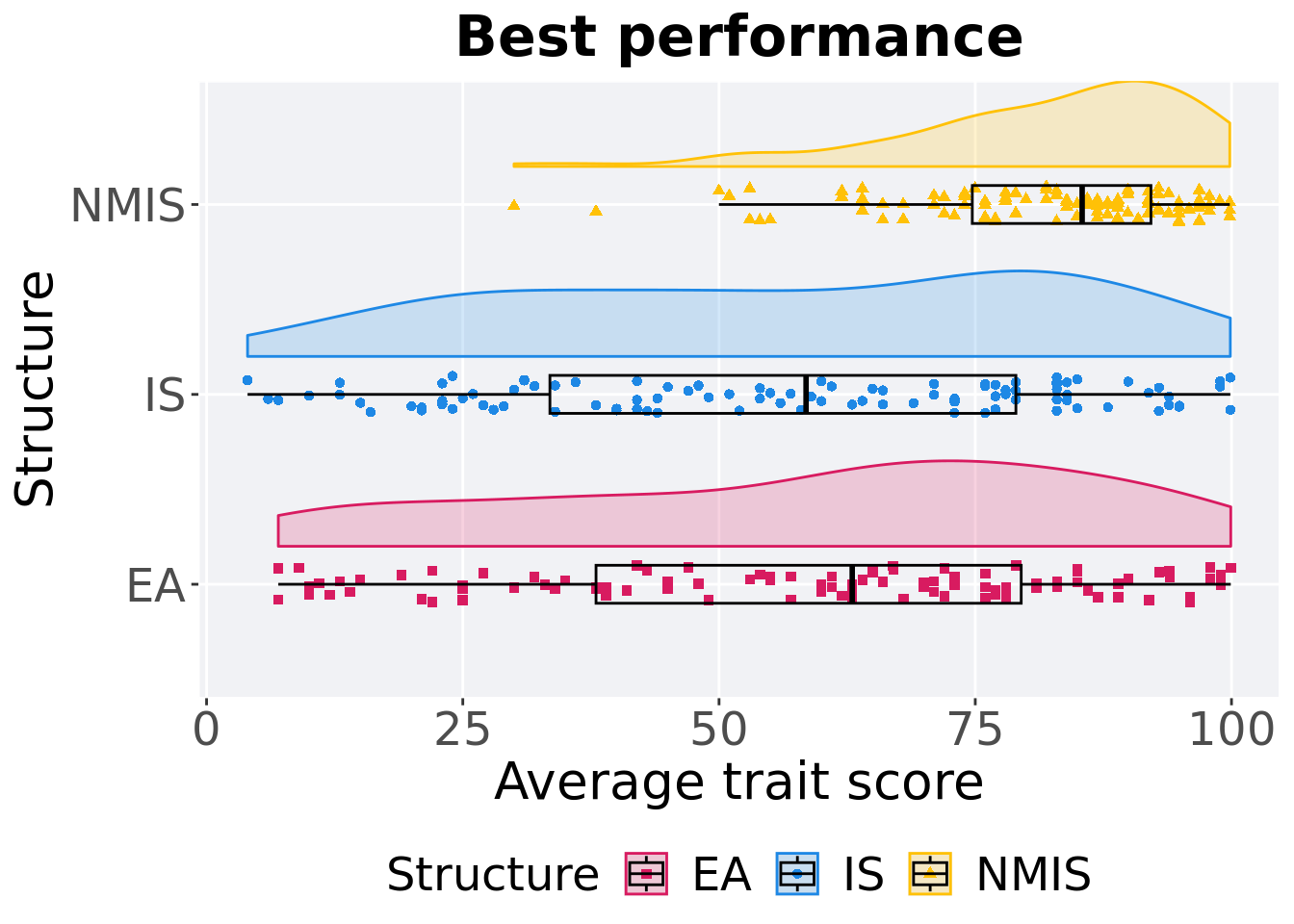
13.3.1.2.1 Stats
Summary statistics for the first generation a satisfactory solution is found.
performance = filter(mi50_best, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TOURNAMENT' & VAR == 'pop_fit_max')
performance %>%
group_by(Structure) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(VAL)),
min = min(VAL, na.rm = TRUE) / DIMENSIONALITY,
median = median(VAL, na.rm = TRUE) / DIMENSIONALITY,
mean = mean(VAL, na.rm = TRUE) / DIMENSIONALITY,
max = max(VAL, na.rm = TRUE) / DIMENSIONALITY,
IQR = IQR(VAL, na.rm = TRUE) / DIMENSIONALITY
)## # A tibble: 3 x 8
## Structure count na_cnt min median mean max IQR
## <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 EA 100 0 7.00 63.0 58.2 99.9 41.5
## 2 IS 100 0 4 58.5 56.7 99.9 45.5
## 3 NMIS 100 0 30.0 85.4 81.7 99.8 17.4Kruskal–Wallis test provides evidence of difference among selection schemes.
##
## Kruskal-Wallis rank sum test
##
## data: VAL by Structure
## Kruskal-Wallis chi-squared = 56.11, df = 2, p-value = 6.546e-13Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction.
pairwise.wilcox.test(x = performance$VAL, g = performance$Structure, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'g')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: performance$VAL and performance$Structure
##
## EA IS
## IS 1 -
## NMIS 4.2e-10 5.1e-11
##
## P value adjustment method: bonferroni13.3.1.3 Final performance
First generation a satisfactory solution is found throughout the 50,000 generations.
filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TOURNAMENT' & Generations == 50000) %>%
ggplot(., aes(x = Structure, y = pop_fit_max / DIMENSIONALITY, color = Structure, fill = Structure, shape = Structure)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_discrete(
name="Structure"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette, ) +
scale_fill_manual(values = cb_palette) +
ggtitle('Final performance')+
p_theme + coord_flip()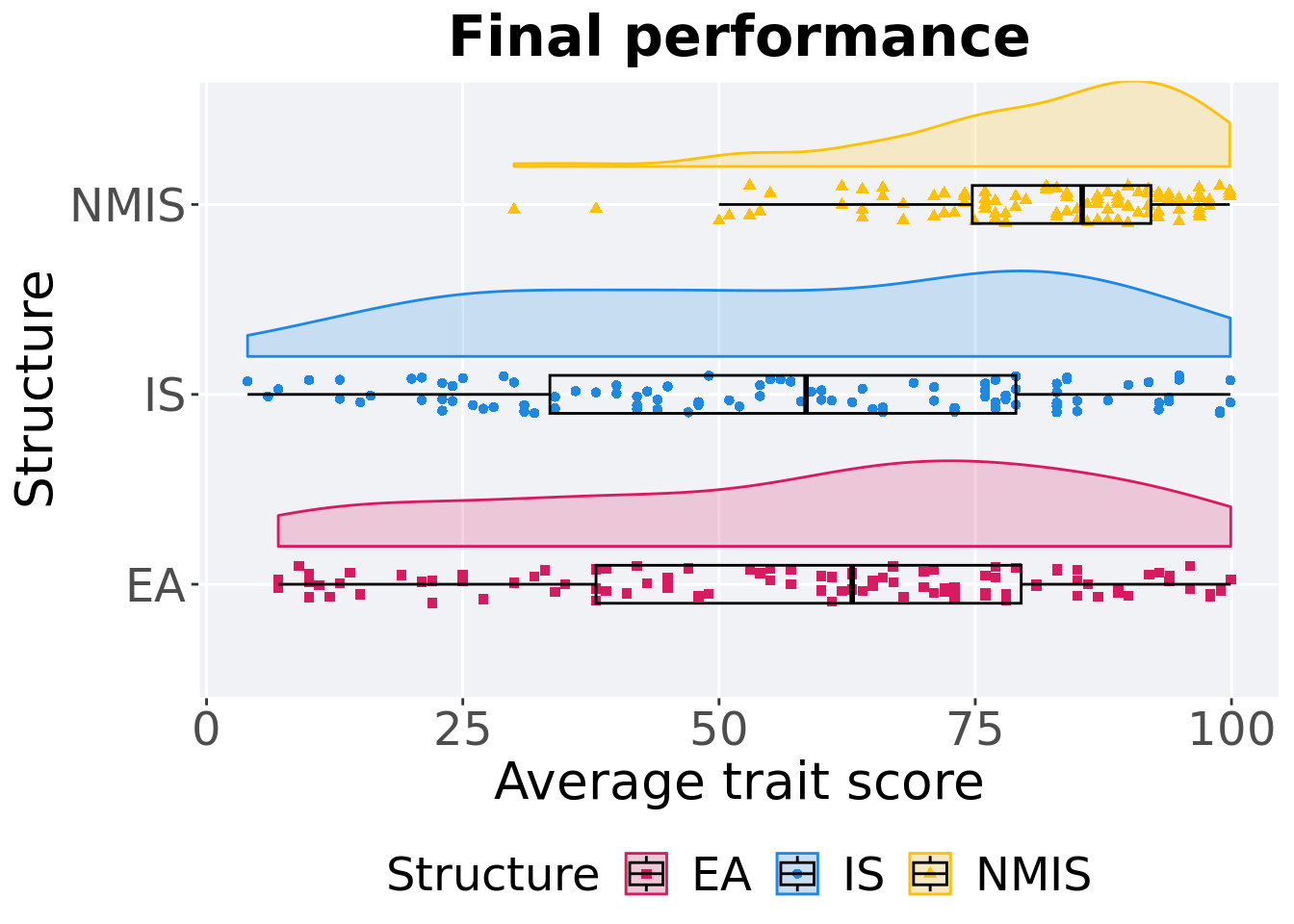
13.3.1.3.1 Stats
Summary statistics for the first generation a satisfactory solution is found.
performance = filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TOURNAMENT' & Generations == 50000)
performance %>%
group_by(Structure) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_fit_max)),
min = min(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
median = median(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
mean = mean(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
max = max(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
IQR = IQR(pop_fit_max / DIMENSIONALITY, na.rm = TRUE)
)## # A tibble: 3 x 8
## Structure count na_cnt min median mean max IQR
## <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 EA 100 0 7.00 63.0 58.2 99.9 41.5
## 2 IS 100 0 4 58.5 56.7 99.9 45.5
## 3 NMIS 100 0 30.0 85.4 81.7 99.8 17.4Kruskal–Wallis test provides evidence of difference among selection schemes.
##
## Kruskal-Wallis rank sum test
##
## data: pop_fit_max by Structure
## Kruskal-Wallis chi-squared = 56.11, df = 2, p-value = 6.546e-13Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction.
pairwise.wilcox.test(x = performance$pop_fit_max, g = performance$Structure, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'g')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: performance$pop_fit_max and performance$Structure
##
## EA IS
## IS 1 -
## NMIS 4.2e-10 5.1e-11
##
## P value adjustment method: bonferroni13.3.2 Generation satisfactory solution found
First generation a satisfactory solution is found throughout the 50,000 generations.
filter(mi50_ssf, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TOURNAMENT'& Generations <= GENERATIONS) %>%
ggplot(., aes(x = Structure, y = Generations, color = Structure, fill = Structure, shape = Structure)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_shape_manual(values=SHAPE)+
scale_y_continuous(
name="Generations"
) +
scale_x_discrete(
name="Structure"
) +
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
p_theme + coord_flip()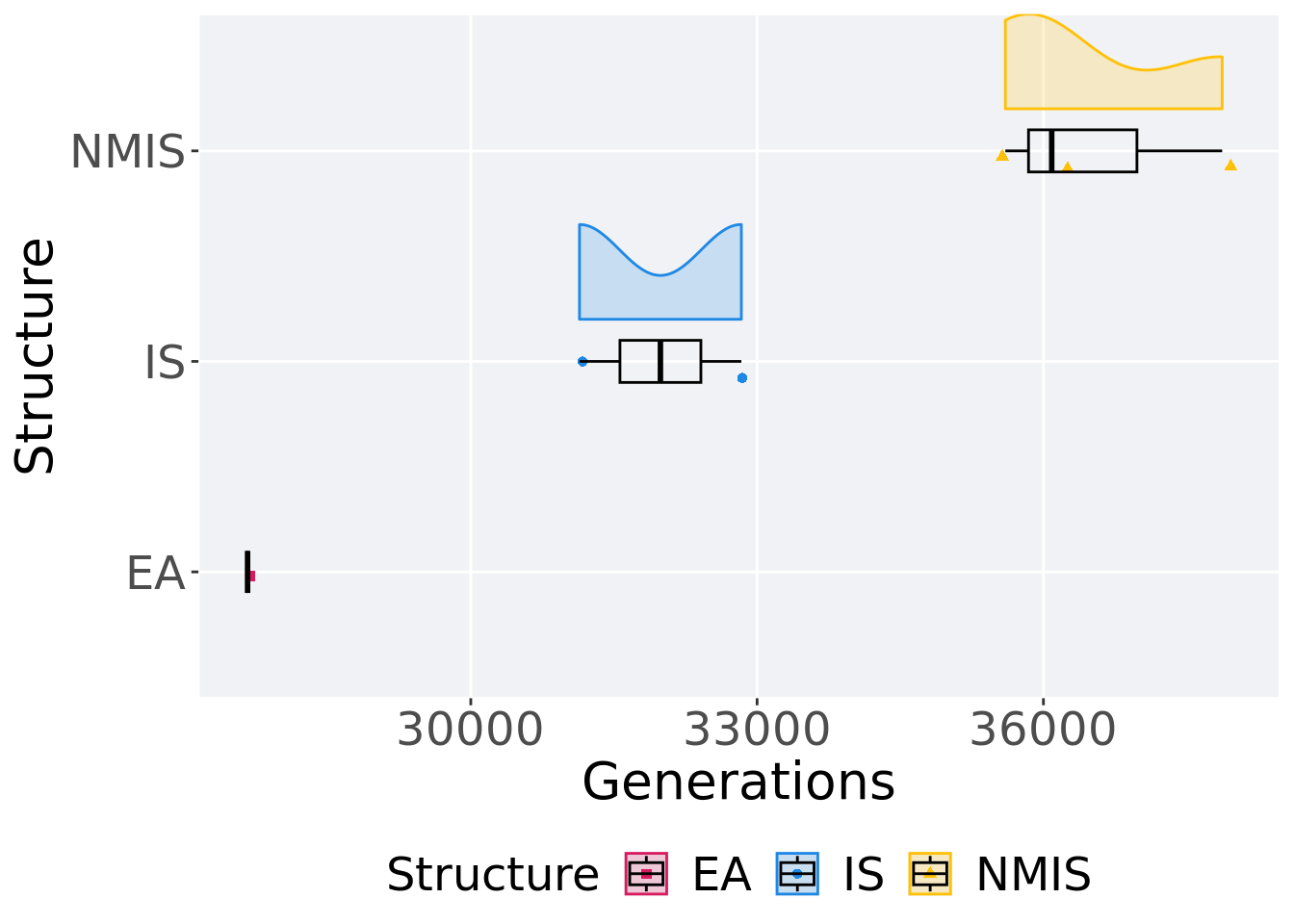
13.3.2.1 Stats
Summary statistics for the first generation a satisfactory solution is found.
ssf = filter(mi50_ssf, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TOURNAMENT' & Generations < 60000)
ssf %>%
group_by(Structure) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(Generations)),
min = min(Generations, na.rm = TRUE),
median = median(Generations, na.rm = TRUE),
mean = mean(Generations, na.rm = TRUE),
max = max(Generations, na.rm = TRUE),
IQR = IQR(Generations, na.rm = TRUE)
)## # A tibble: 3 x 8
## Structure count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 EA 1 0 27661 27661 27661 27661 0
## 2 IS 2 0 31139 31987 31987 32835 848
## 3 NMIS 3 0 35601 36087 36520. 37873 1136Kruskal–Wallis test provides evidence of no difference among selection schemes.
##
## Kruskal-Wallis rank sum test
##
## data: Generations by Structure
## Kruskal-Wallis chi-squared = 4.2857, df = 2, p-value = 0.1173pairwise.wilcox.test(x = ssf$Generations, g = ssf$Structure, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'g')##
## Pairwise comparisons using Wilcoxon rank sum exact test
##
## data: ssf$Generations and ssf$Structure
##
## EA IS
## IS 1.00 -
## NMIS 0.75 0.30
##
## P value adjustment method: bonferroni13.3.3 Activation gene coverage
Activation gene coverage analysis.
13.3.3.1 Coverage over time
Activation gene coverage over time.
# data for lines and shading on plots
lines = filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TOURNAMENT') %>%
group_by(Structure, Generations) %>%
dplyr::summarise(
min = min(pop_act_cov),
mean = mean(pop_act_cov),
max = max(pop_act_cov)
)## `summarise()` has grouped output by 'Structure'. You can override using the
## `.groups` argument.ggplot(lines, aes(x=Generations, y=mean, group = Structure, fill = Structure, color = Structure, shape = Structure)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, Generations %% 2000 == 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Activation gene coverage over time')+
p_theme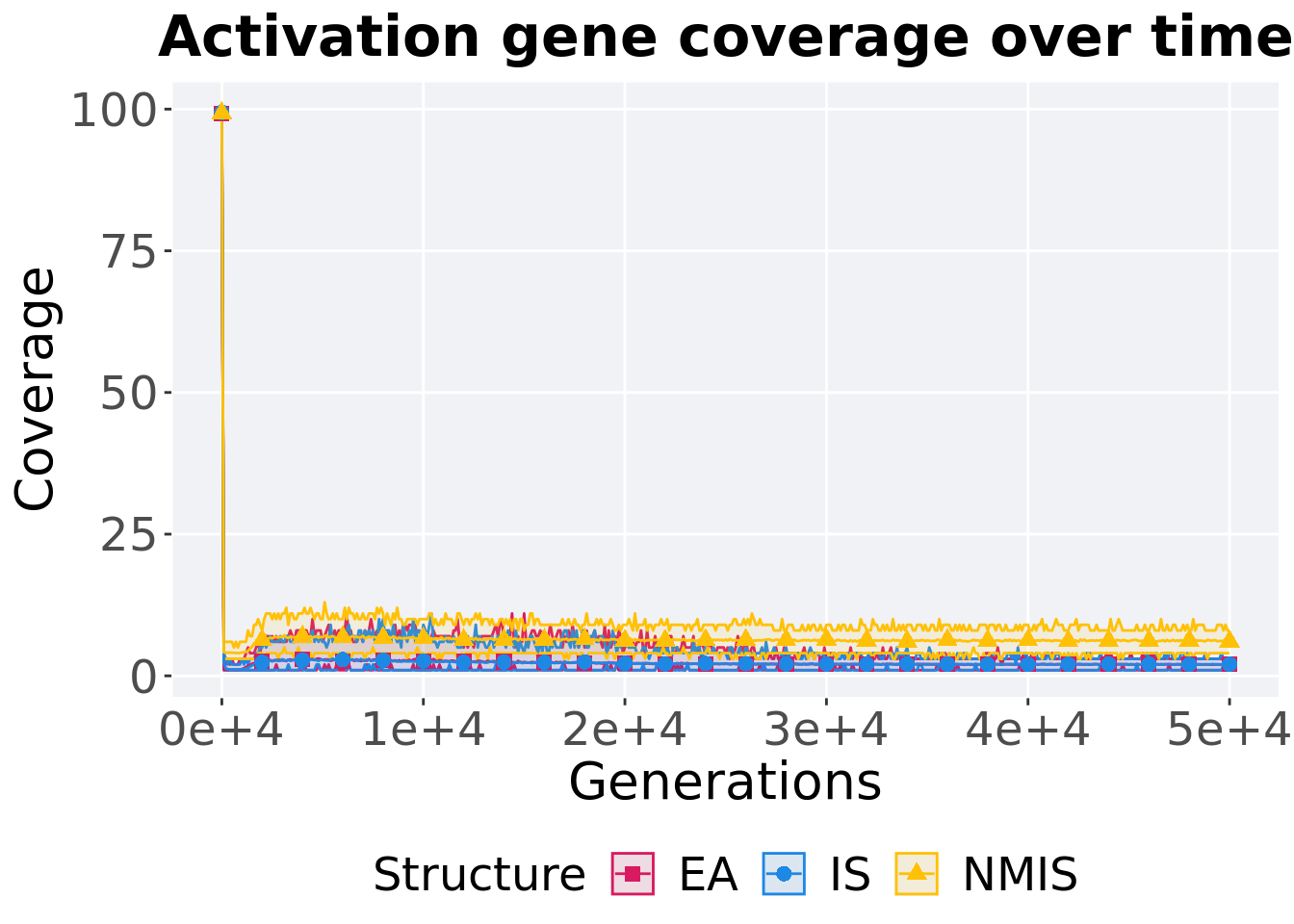
13.3.3.2 End of 50,000 generations
Activation gene coverage in the population at the end of 50,000 generations.
### end of run
filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TOURNAMENT' & Generations == 50000) %>%
ggplot(., aes(x = Structure, y = pop_act_cov, color = Structure, fill = Structure, shape = Structure)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.3) +
geom_point(position = position_jitter(height = .05, width = .05), size = 1.5, alpha = 0.5) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_shape_manual(values=SHAPE)+
scale_y_continuous(
name="Coverage"
) +
scale_x_discrete(
name="Structure"
) +
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Final activation gene coverage')+
p_theme + coord_flip()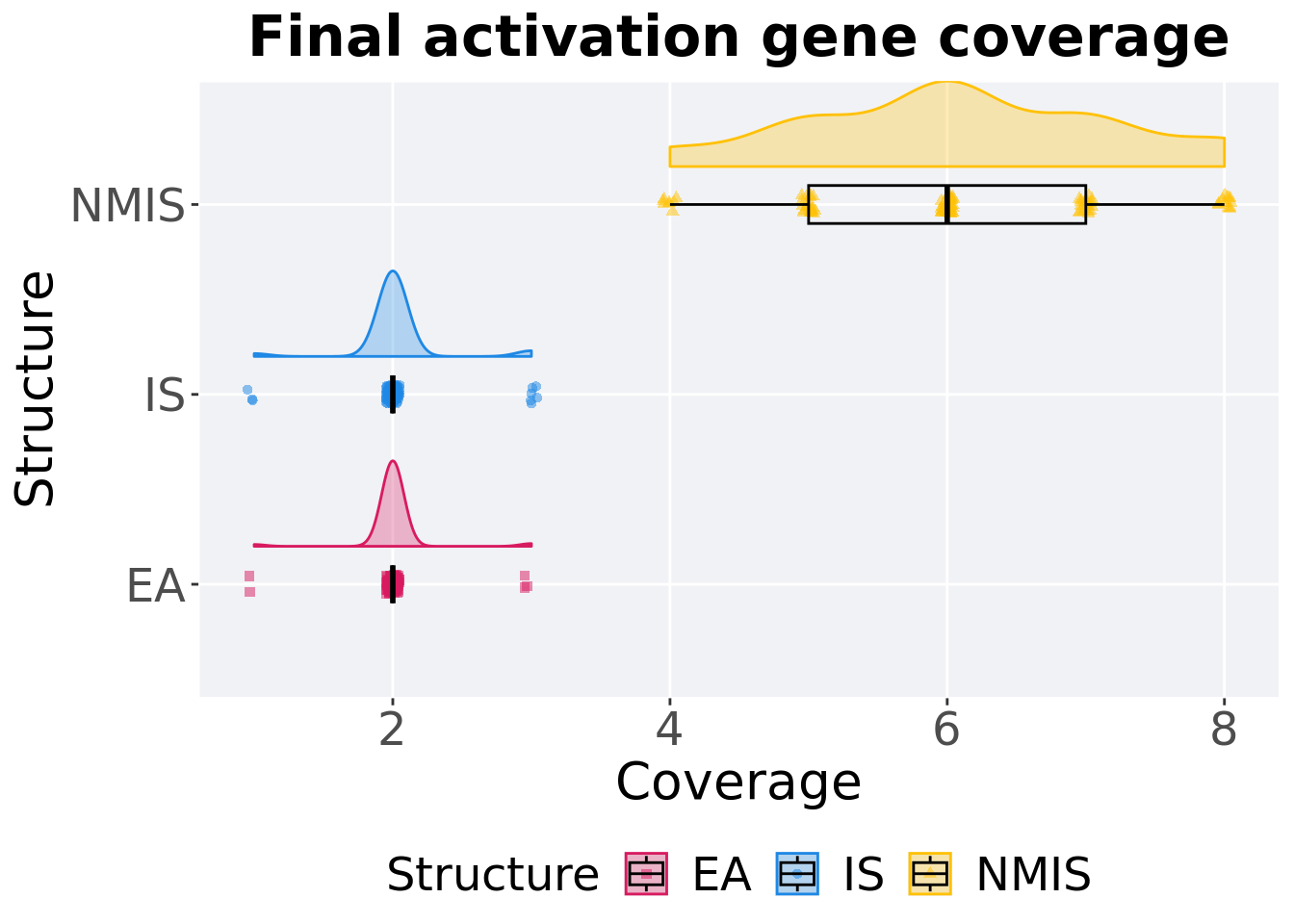
13.3.3.2.1 Stats
Summary statistics for activation gene coverage.
coverage = filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'TOURNAMENT' & Generations == 50000)
coverage %>%
group_by(Structure) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_act_cov)),
min = min(pop_act_cov, na.rm = TRUE),
median = median(pop_act_cov, na.rm = TRUE),
mean = mean(pop_act_cov, na.rm = TRUE),
max = max(pop_act_cov, na.rm = TRUE),
IQR = IQR(pop_act_cov, na.rm = TRUE)
)## # A tibble: 3 x 8
## Structure count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 EA 100 0 1 2 2.01 3 0
## 2 IS 100 0 1 2 2.03 3 0
## 3 NMIS 100 0 4 6 6.09 8 2Kruskal–Wallis test provides evidence of difference among activation gene coverage.
##
## Kruskal-Wallis rank sum test
##
## data: pop_act_cov by Structure
## Kruskal-Wallis chi-squared = 262.68, df = 2, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction on activation gene coverage.
pairwise.wilcox.test(x = coverage$pop_act_cov, g = coverage$Structure, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'g')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: coverage$pop_act_cov and coverage$Structure
##
## EA IS
## IS 0.88 -
## NMIS <2e-16 <2e-16
##
## P value adjustment method: bonferroni13.4 Lexicase selection
Here we analyze how the different population structures affect standard lexicase selection on the contradictory objectives diagnostic.
13.4.1 Performance
13.4.1.1 Performance over time
lines = filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'LEXICASE') %>%
group_by(Structure, Generations) %>%
dplyr::summarise(
min = min(pop_fit_max) / DIMENSIONALITY,
mean = mean(pop_fit_max) / DIMENSIONALITY,
max = max(pop_fit_max) / DIMENSIONALITY
)
ggplot(lines, aes(x=Generations, y=mean, group = Structure, fill = Structure, color = Structure, shape = Structure)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, Generations %% 2000 == 0), size = 2.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Performance over time") +
p_theme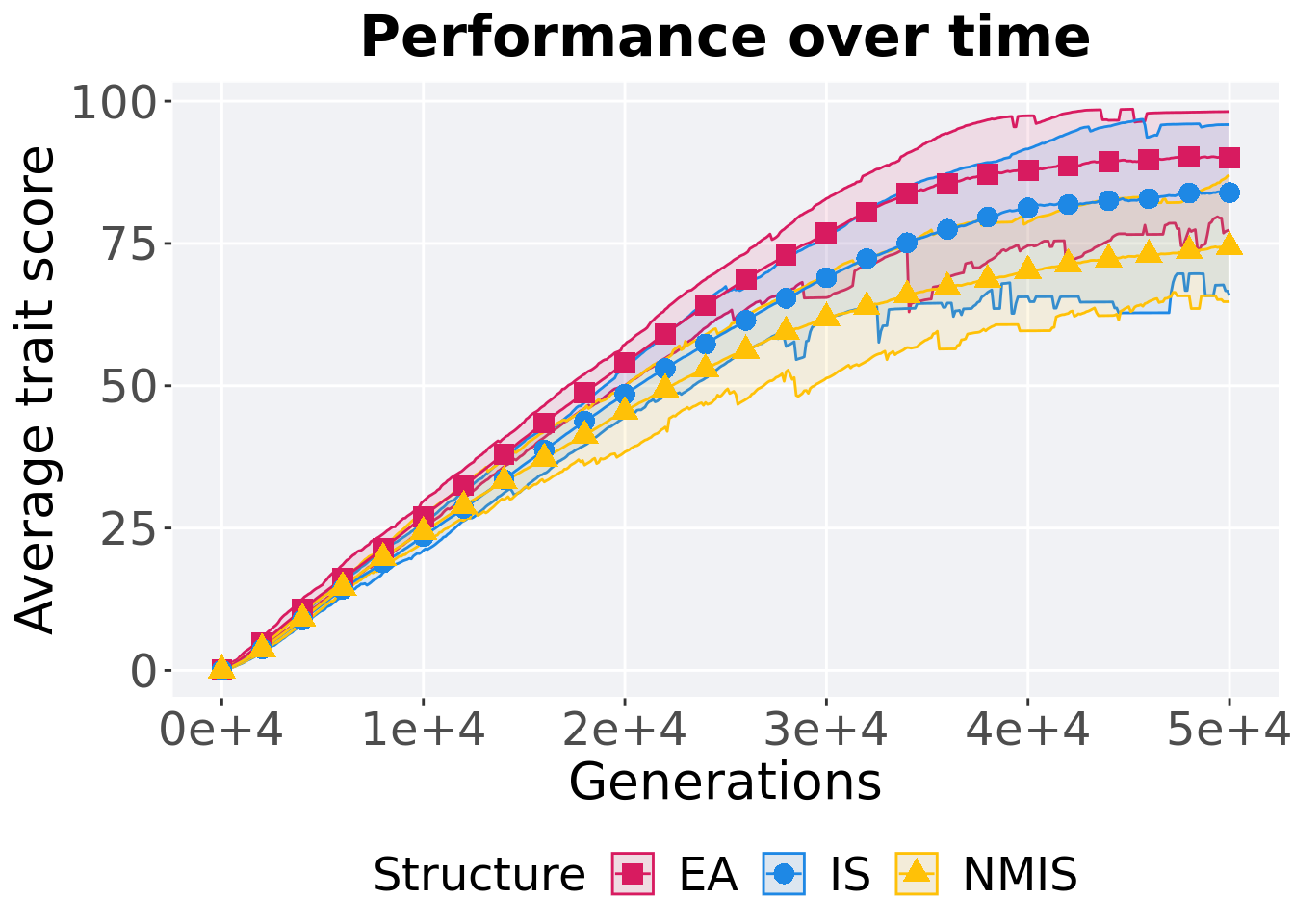
13.4.1.2 Best performance
First generation a satisfactory solution is found throughout the 50,000 generations.
filter(mi50_best, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'LEXICASE' & VAR == 'pop_fit_max') %>%
ggplot(., aes(x = Structure, y = VAL / DIMENSIONALITY, color = Structure, fill = Structure, shape = Structure)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_discrete(
name="Structure"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette, ) +
scale_fill_manual(values = cb_palette) +
ggtitle('Best performance')+
p_theme + coord_flip()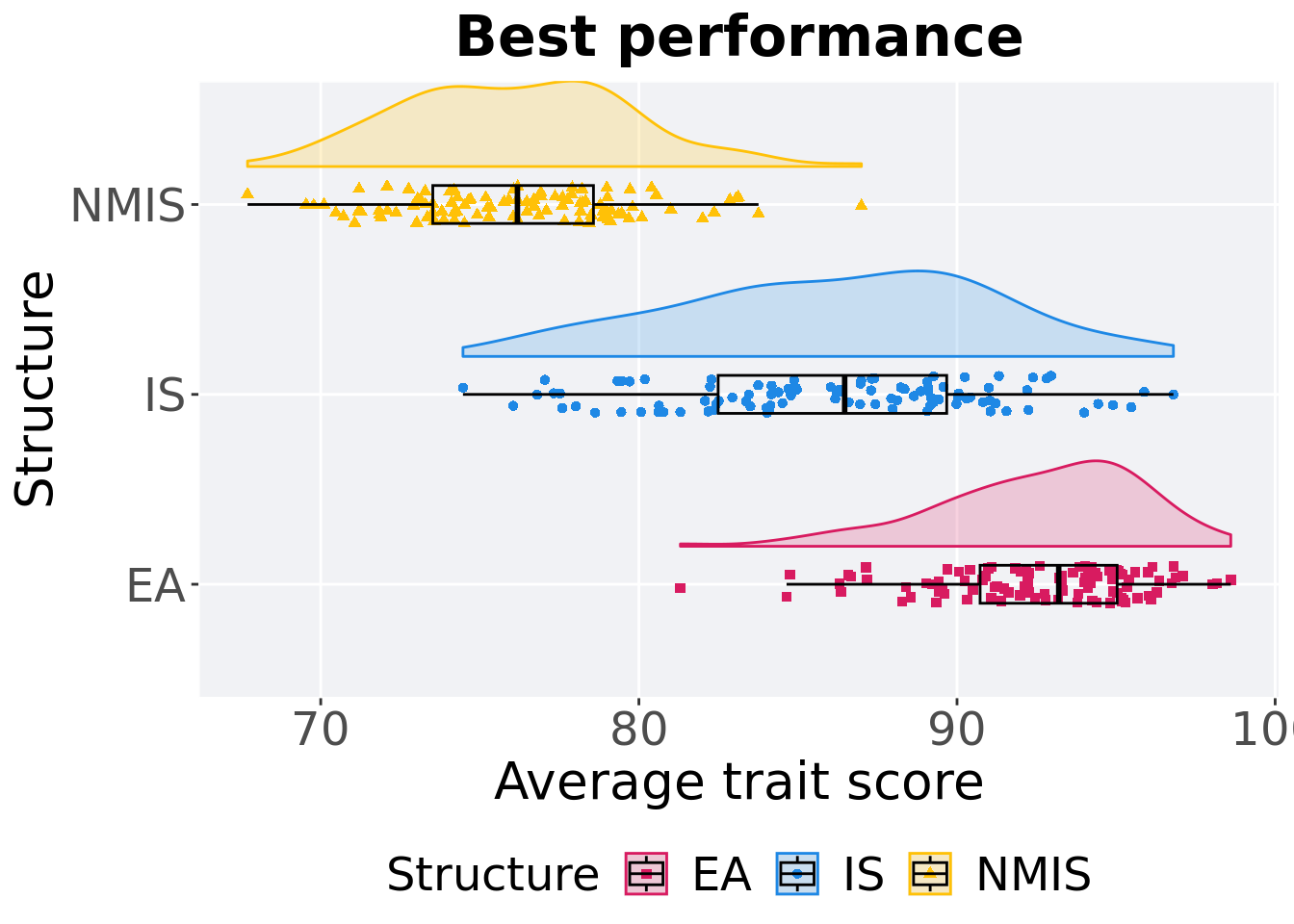
13.4.1.2.1 Stats
Summary statistics for the first generation a satisfactory solution is found.
performance = filter(mi50_best, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'LEXICASE' & VAR == 'pop_fit_max')
performance$Structure = factor(performance$Structure, levels=c('EA','NMIS','IS'))
performance %>%
group_by(Structure) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(VAL)),
min = min(VAL, na.rm = TRUE) / DIMENSIONALITY,
median = median(VAL, na.rm = TRUE) / DIMENSIONALITY,
mean = mean(VAL, na.rm = TRUE) / DIMENSIONALITY,
max = max(VAL, na.rm = TRUE) / DIMENSIONALITY,
IQR = IQR(VAL, na.rm = TRUE) / DIMENSIONALITY
)## # A tibble: 3 x 8
## Structure count na_cnt min median mean max IQR
## <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 EA 100 0 81.3 93.2 92.6 98.6 4.31
## 2 NMIS 100 0 67.7 76.2 76.1 87.0 5.05
## 3 IS 100 0 74.5 86.5 86.1 96.8 7.18Kruskal–Wallis test provides evidence of difference among selection schemes.
##
## Kruskal-Wallis rank sum test
##
## data: VAL by Structure
## Kruskal-Wallis chi-squared = 217.42, df = 2, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction.
pairwise.wilcox.test(x = performance$VAL, g = performance$Structure, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'l')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: performance$VAL and performance$Structure
##
## EA NMIS
## NMIS <2e-16 -
## IS <2e-16 1
##
## P value adjustment method: bonferroni13.4.1.3 Final performance
First generation a satisfactory solution is found throughout the 50,000 generations.
filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'LEXICASE' & Generations == 50000) %>%
ggplot(., aes(x = Structure, y = pop_fit_max / DIMENSIONALITY, color = Structure, fill = Structure, shape = Structure)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_discrete(
name="Structure"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette, ) +
scale_fill_manual(values = cb_palette) +
ggtitle('Final performance')+
p_theme + coord_flip()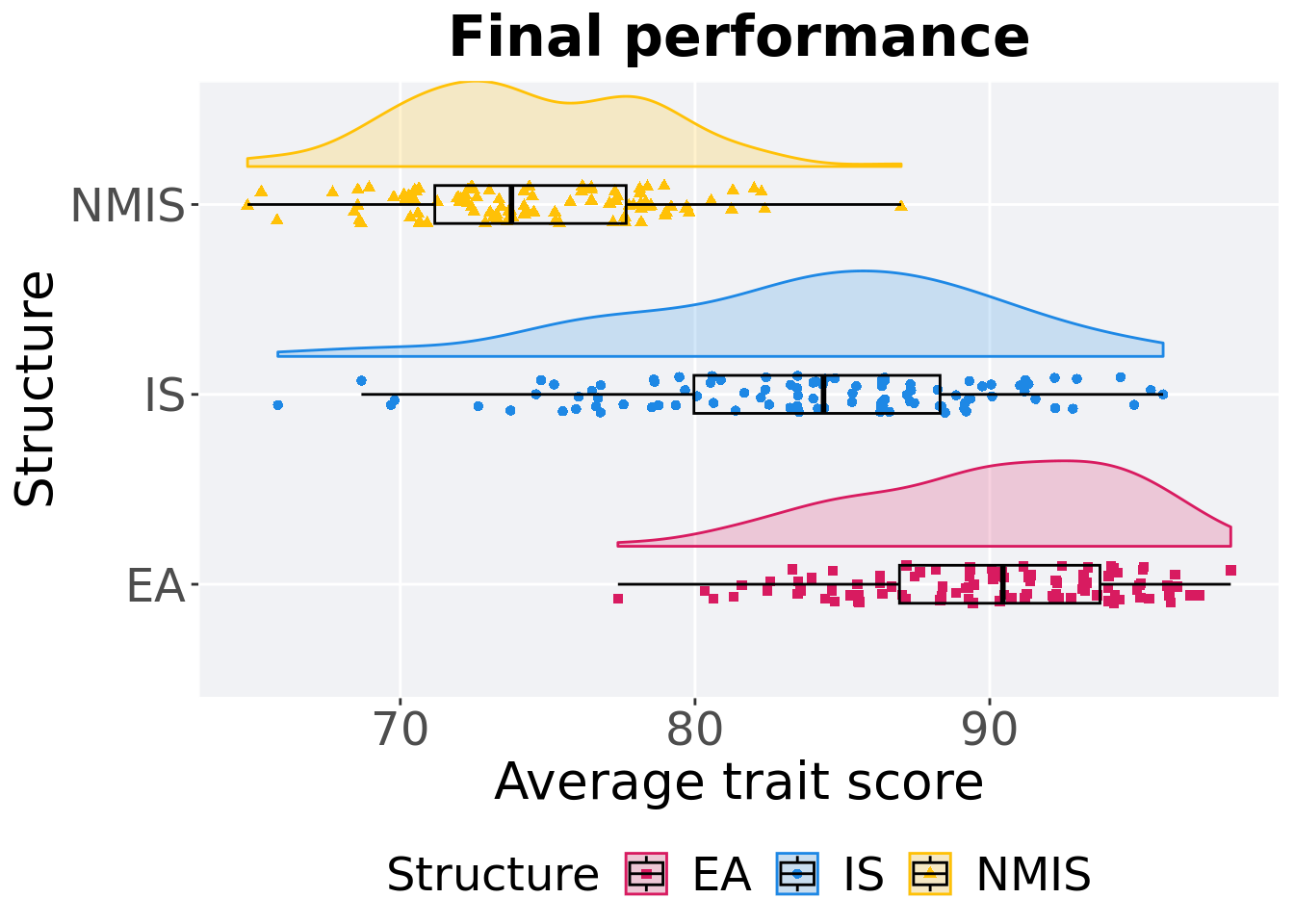
13.4.1.3.1 Stats
Summary statistics for the first generation a satisfactory solution is found.
performance = filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'LEXICASE' & Generations == 50000)
performance$Structure = factor(performance$Structure, levels=c('EA','NMIS','IS'))
performance %>%
group_by(Structure) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_fit_max)),
min = min(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
median = median(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
mean = mean(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
max = max(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
IQR = IQR(pop_fit_max / DIMENSIONALITY, na.rm = TRUE)
)## # A tibble: 3 x 8
## Structure count na_cnt min median mean max IQR
## <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 EA 100 0 77.4 90.4 90.1 98.2 6.80
## 2 NMIS 100 0 64.8 73.8 74.4 87.0 6.49
## 3 IS 100 0 65.8 84.4 83.9 95.9 8.35Kruskal–Wallis test provides evidence of difference among selection schemes.
##
## Kruskal-Wallis rank sum test
##
## data: pop_fit_max by Structure
## Kruskal-Wallis chi-squared = 186.92, df = 2, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction.
pairwise.wilcox.test(x = performance$pop_fit_max, g = performance$Structure, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'l')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: performance$pop_fit_max and performance$Structure
##
## EA NMIS
## NMIS < 2e-16 -
## IS 2.5e-12 1
##
## P value adjustment method: bonferroni13.4.2 Activation gene coverage
Activation gene coverage analysis.
13.4.2.1 Coverage over time
Activation gene coverage over time.
# data for lines and shading on plots
lines = filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'LEXICASE') %>%
group_by(Structure, Generations) %>%
dplyr::summarise(
min = min(pop_act_cov),
mean = mean(pop_act_cov),
max = max(pop_act_cov)
)## `summarise()` has grouped output by 'Structure'. You can override using the
## `.groups` argument.ggplot(lines, aes(x=Generations, y=mean, group = Structure, fill = Structure, color = Structure, shape = Structure)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, Generations %% 2000 == 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Activation gene coverage over time')+
p_theme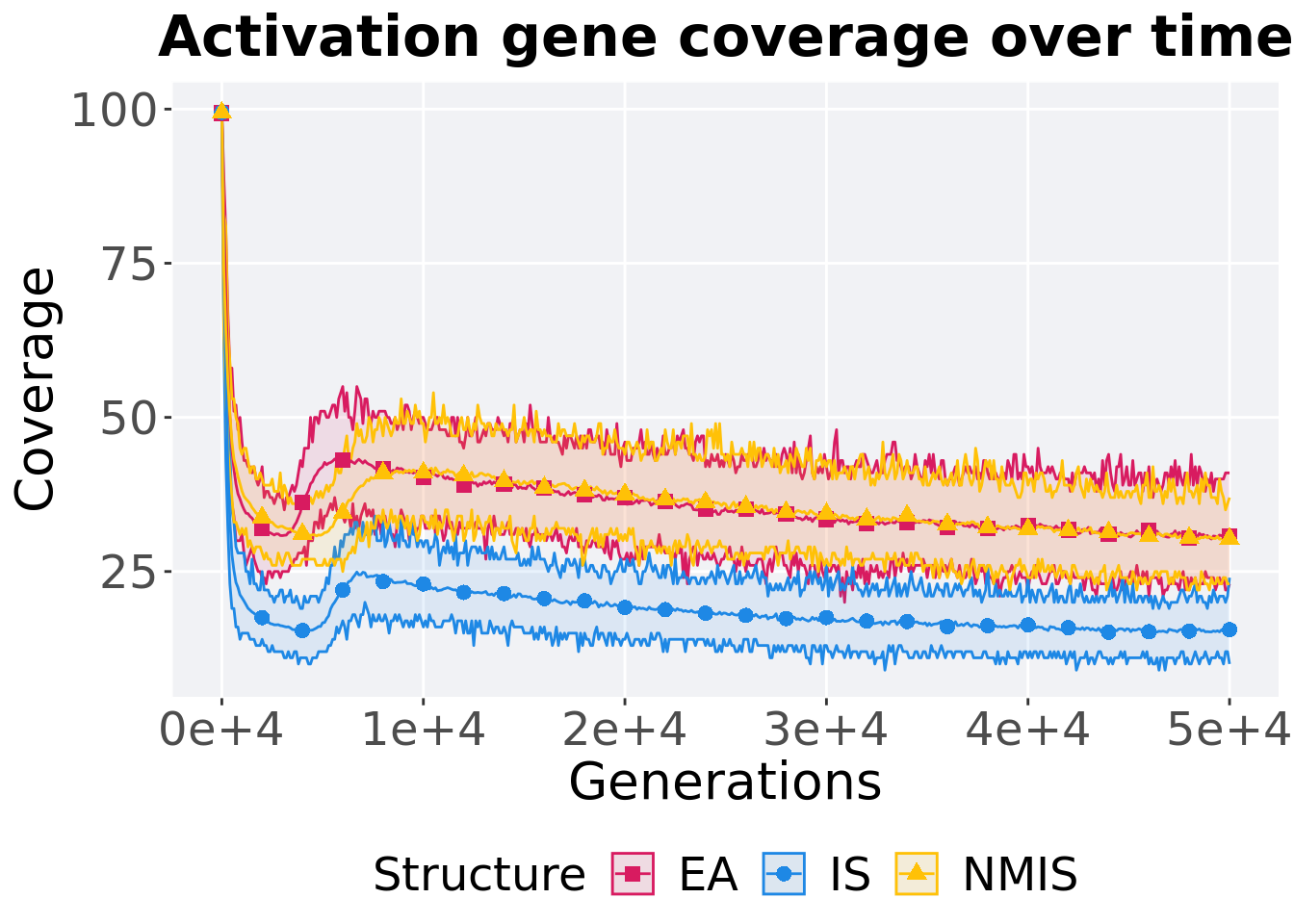
13.4.2.2 End of 50,000 generations
Activation gene coverage in the population at the end of 50,000 generations.
### end of run
filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'LEXICASE' & Generations == 50000) %>%
ggplot(., aes(x = Structure, y = pop_act_cov, color = Structure, fill = Structure, shape = Structure)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.3) +
geom_point(position = position_jitter(height = .05, width = .05), size = 1.5, alpha = 0.5) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_shape_manual(values=SHAPE)+
scale_y_continuous(
name="Coverage"
) +
scale_x_discrete(
name="Structure"
) +
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Final activation gene coverage')+
p_theme + coord_flip()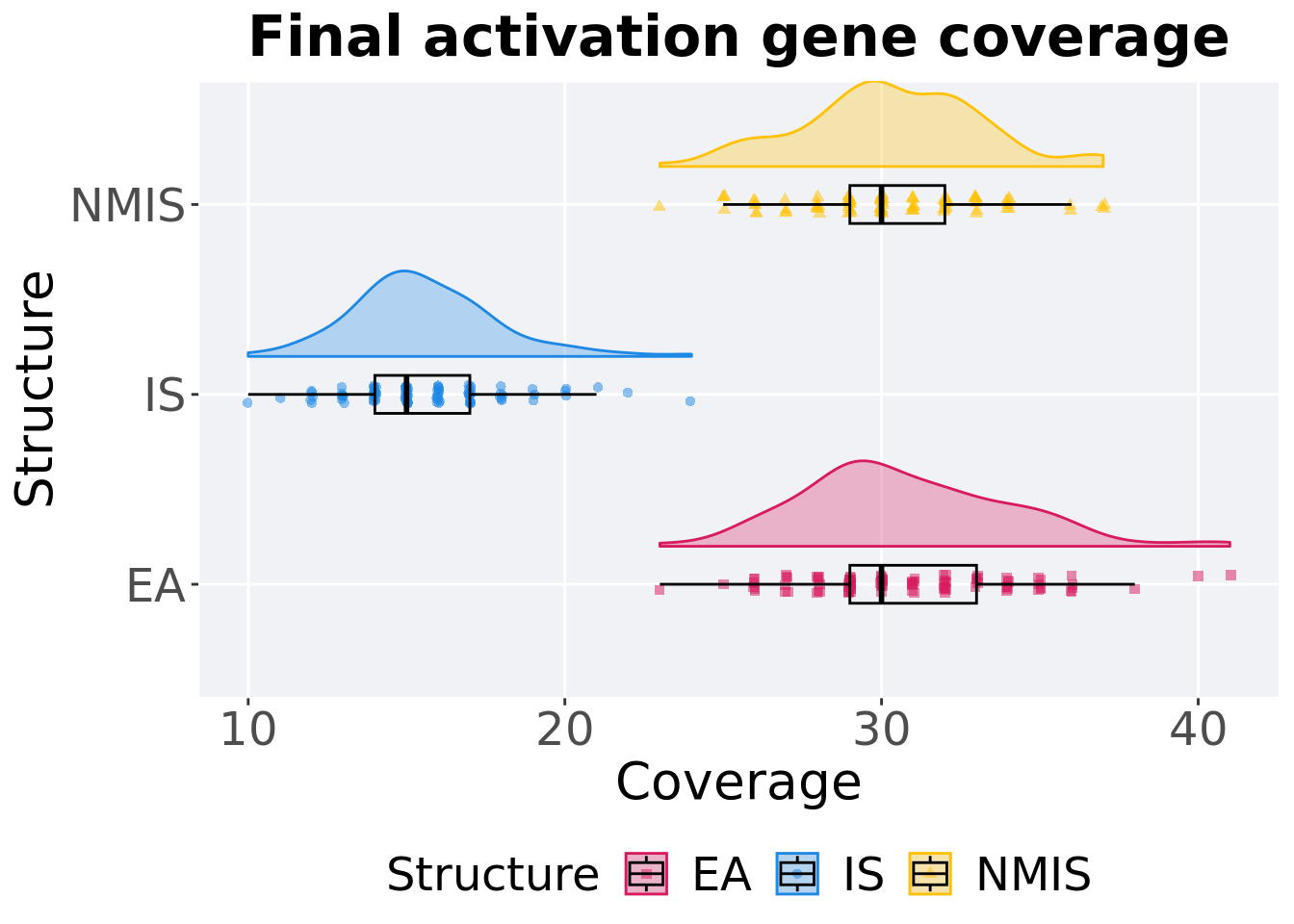
13.4.2.2.1 Stats
Summary statistics for activation gene coverage.
coverage = filter(mi50_over_time, Diagnostic == 'MULTIPATH_EXPLORATION' & `Selection\nScheme` == 'LEXICASE' & Generations == 50000)
coverage$Structure = factor(coverage$Structure, levels=c('EA','NMIS','IS'))
coverage %>%
group_by(Structure) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_act_cov)),
min = min(pop_act_cov, na.rm = TRUE),
median = median(pop_act_cov, na.rm = TRUE),
mean = mean(pop_act_cov, na.rm = TRUE),
max = max(pop_act_cov, na.rm = TRUE),
IQR = IQR(pop_act_cov, na.rm = TRUE)
)## # A tibble: 3 x 8
## Structure count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 EA 100 0 23 30 30.8 41 4
## 2 NMIS 100 0 23 30 30.3 37 3
## 3 IS 100 0 10 15 15.6 24 3Kruskal–Wallis test provides evidence of difference among activation gene coverage.
##
## Kruskal-Wallis rank sum test
##
## data: pop_act_cov by Structure
## Kruskal-Wallis chi-squared = 200.36, df = 2, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction on activation gene coverage.
pairwise.wilcox.test(x = coverage$pop_act_cov, g = coverage$Structure, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'l')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: coverage$pop_act_cov and coverage$Structure
##
## EA NMIS
## NMIS 0.81 -
## IS <2e-16 <2e-16
##
## P value adjustment method: bonferroni