Chapter 7 Tournament selection
We present the results from our parameter sweeep on tournament selection.
50 replicates are conducted for each tournament size T parameter value explored.
7.1 Exploitation rate results
Here we present the results for best performances found by each tournament selection value replicate on the exploitation rate diagnostic.
7.1.1 Performance over time
Performance over time.
lines = filter(tor_ot, diagnostic == 'exploitation_rate') %>%
group_by(T, gen) %>%
dplyr::summarise(
min = min(pop_fit_max),
mean = mean(pop_fit_max),
max = max(pop_fit_max)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.ggplot(lines, aes(x=gen, y=mean / DIMENSIONALITY, group = T, fill = T, color = T, shape = T)) +
geom_ribbon(aes(ymin = min / DIMENSIONALITY, ymax = max / DIMENSIONALITY), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Best performance over time") +
p_theme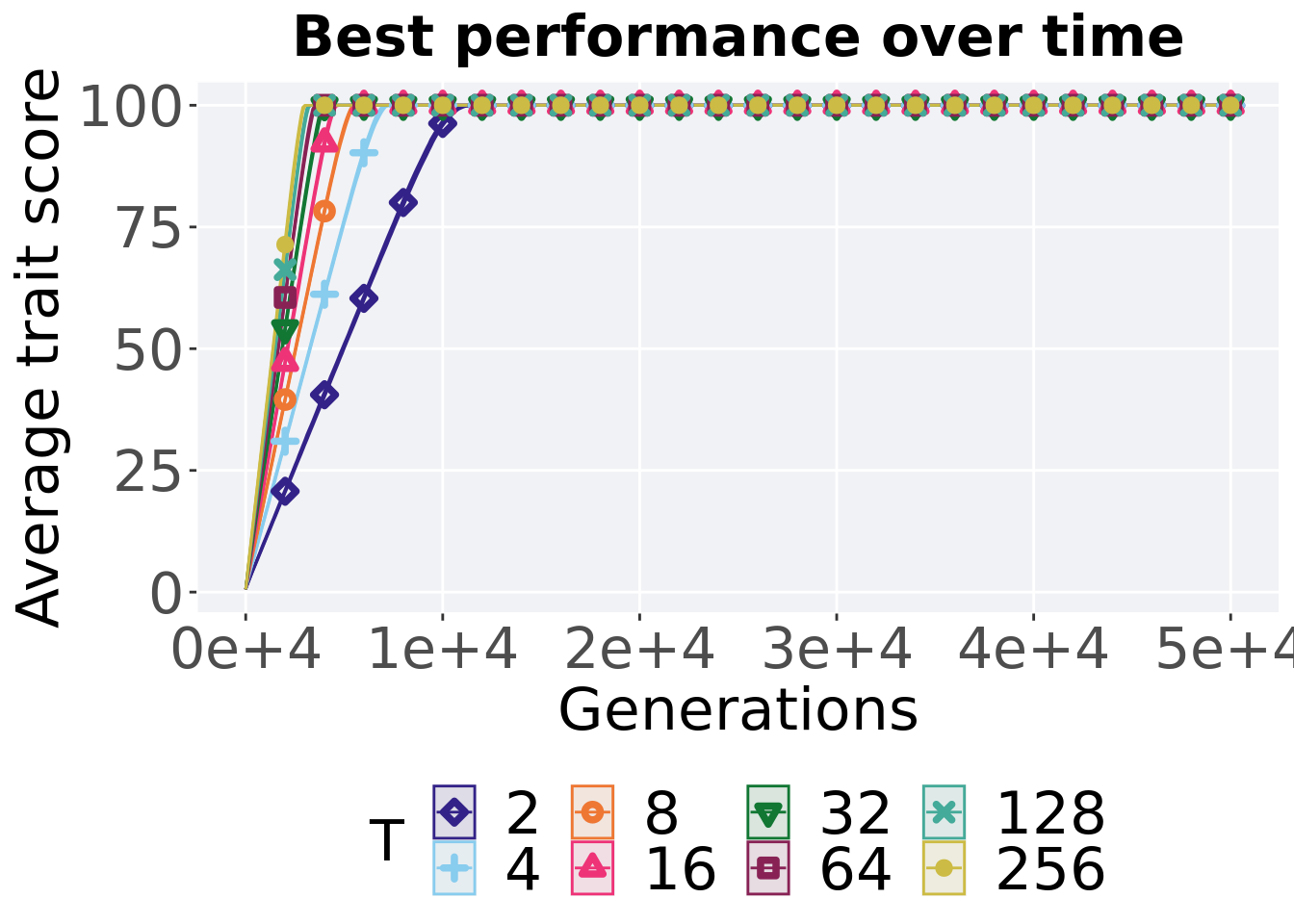
7.1.2 Generation satisfactory solution found
The first Generations a satisfactory solution is found throughout the 50,000 generations.
filter(tor_ssf, Diagnostic == 'EXPLOITATION_RATE') %>%
ggplot(., aes(x = T, y = Generations, color = T, fill = T, shape = T)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_shape_manual(values=SHAPE)+
scale_y_continuous(
name="Generation",
limits=c(0, 12000),
breaks=c(0, 2000, 4000, 6000, 8000, 10000, 12000),
labels=c("0e+4", "2e+4", "4e+4", "6e+4", "8e+4", "10e+4", "12e+4")
) +
scale_x_discrete(
name="T"
) +
ggtitle("Generation satisfactory solution found") +
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
p_theme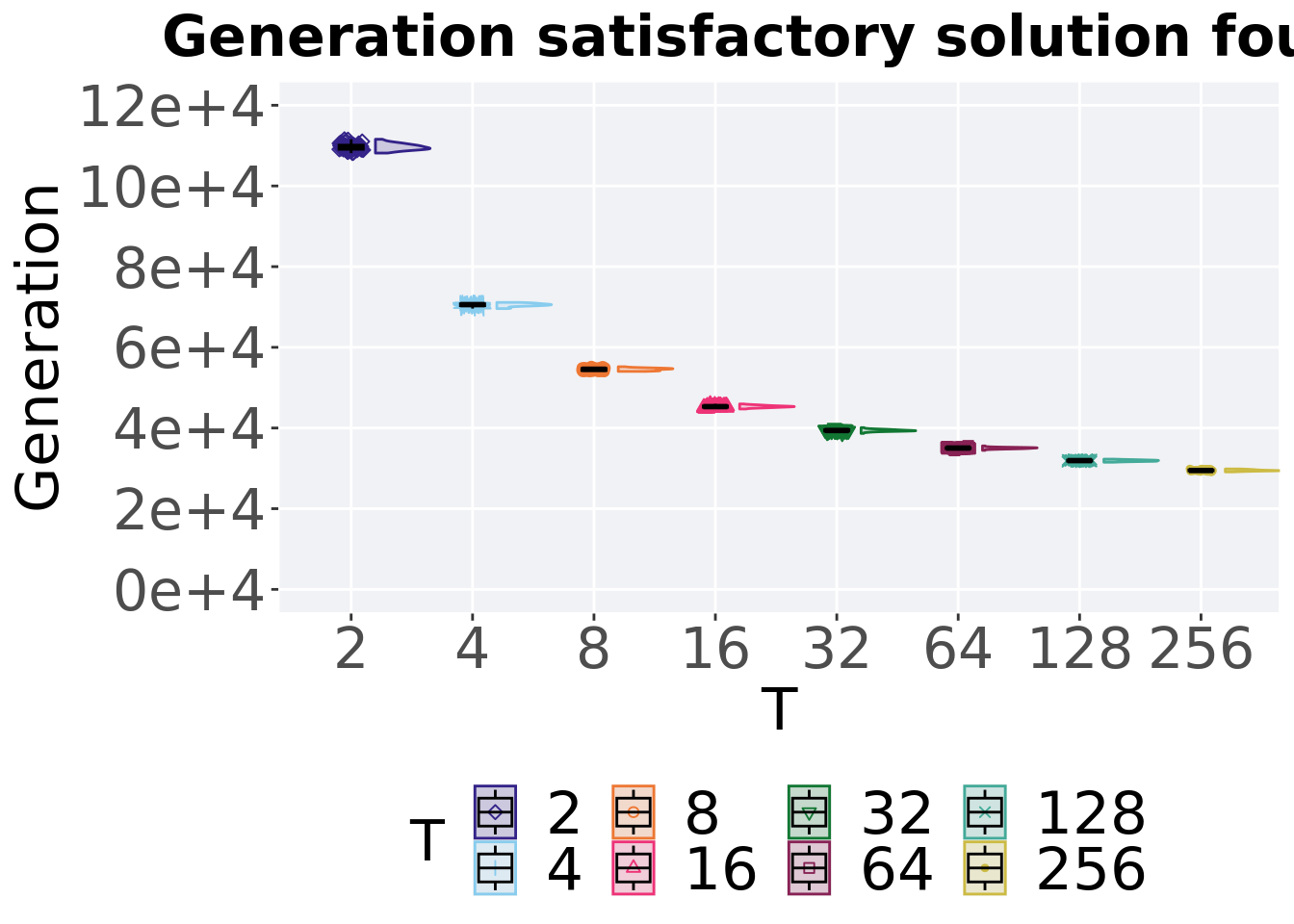
7.1.2.1 Stats
Summary statistics for the best performance found throughout 50,000 generations.
ssf = filter(tor_ssf, Diagnostic == 'EXPLOITATION_RATE')
group_by(ssf, T) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(Generations)),
min = min(Generations, na.rm = TRUE),
median = median(Generations, na.rm = TRUE),
mean = mean(Generations, na.rm = TRUE),
max = max(Generations, na.rm = TRUE),
IQR = IQR(Generations, na.rm = TRUE)
)## # A tibble: 8 x 8
## T count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 2 50 0 10815 10955 10966. 11157 102.
## 2 4 50 0 6960 7059 7052. 7113 48.8
## 3 8 50 0 5403 5457 5453. 5519 51.8
## 4 16 50 0 4471 4530. 4532. 4597 33.8
## 5 32 50 0 3865 3938 3939. 4009 31.8
## 6 64 50 0 3446 3504. 3502. 3556 24.2
## 7 128 50 0 3152 3192 3190. 3228 26.8
## 8 256 50 0 2912 2946. 2950. 2985 24.8Kruskal–Wallis test provides evidence of significant differences among the Generations a satisfactory solution is first found.
##
## Kruskal-Wallis rank sum test
##
## data: Generations by T
## Kruskal-Wallis chi-squared = 392.77, df = 7, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction on the Generations a satisfactory solution is first found. .
pairwise.wilcox.test(x = ssf$Generations, g = ssf$T , p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'l')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: ssf$Generations and ssf$T
##
## 2 4 8 16 32 64 128
## 4 <2e-16 - - - - - -
## 8 <2e-16 <2e-16 - - - - -
## 16 <2e-16 <2e-16 <2e-16 - - - -
## 32 <2e-16 <2e-16 <2e-16 <2e-16 - - -
## 64 <2e-16 <2e-16 <2e-16 <2e-16 <2e-16 - -
## 128 <2e-16 <2e-16 <2e-16 <2e-16 <2e-16 <2e-16 -
## 256 <2e-16 <2e-16 <2e-16 <2e-16 <2e-16 <2e-16 <2e-16
##
## P value adjustment method: bonferroni7.1.3 Multi-valley crossing
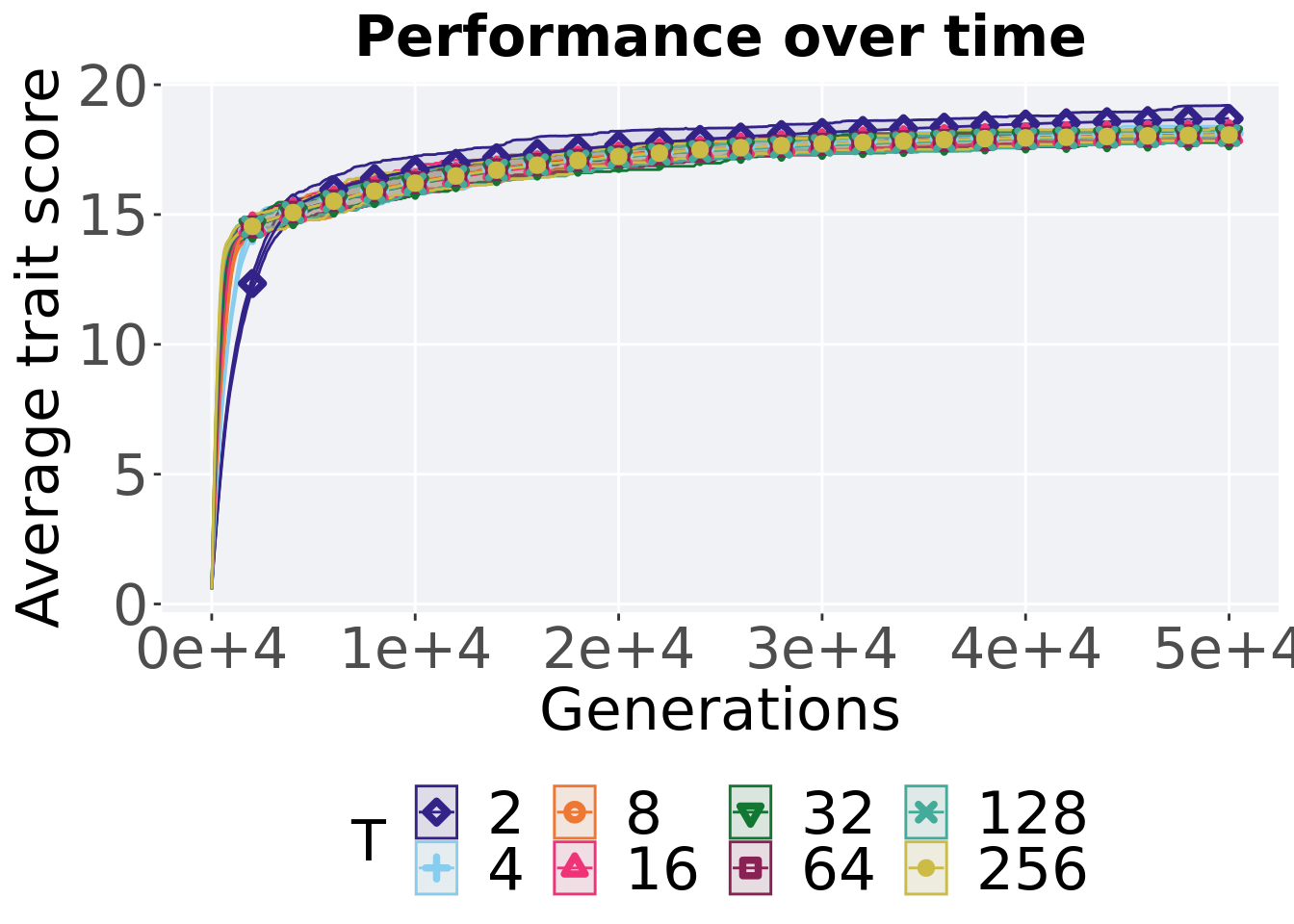
7.1.3.1 Performance over time
# data for lines and shading on plots
lines = filter(tor_ot_mvc, diagnostic == 'exploitation_rate') %>%
group_by(T, gen) %>%
dplyr::summarise(
min = min(pop_fit_max) / DIMENSIONALITY,
mean = mean(pop_fit_max) / DIMENSIONALITY,
max = max(pop_fit_max) / DIMENSIONALITY
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.ggplot(lines, aes(x=gen, y=mean, group = T, fill =T, color = T, shape = T)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Performance over time')+
p_theme +
guides(
shape=guide_legend(nrow=2, title.position = "left"),
color=guide_legend(nrow=2, title.position = "left"),
fill=guide_legend(nrow=2, title.position = "left")
)
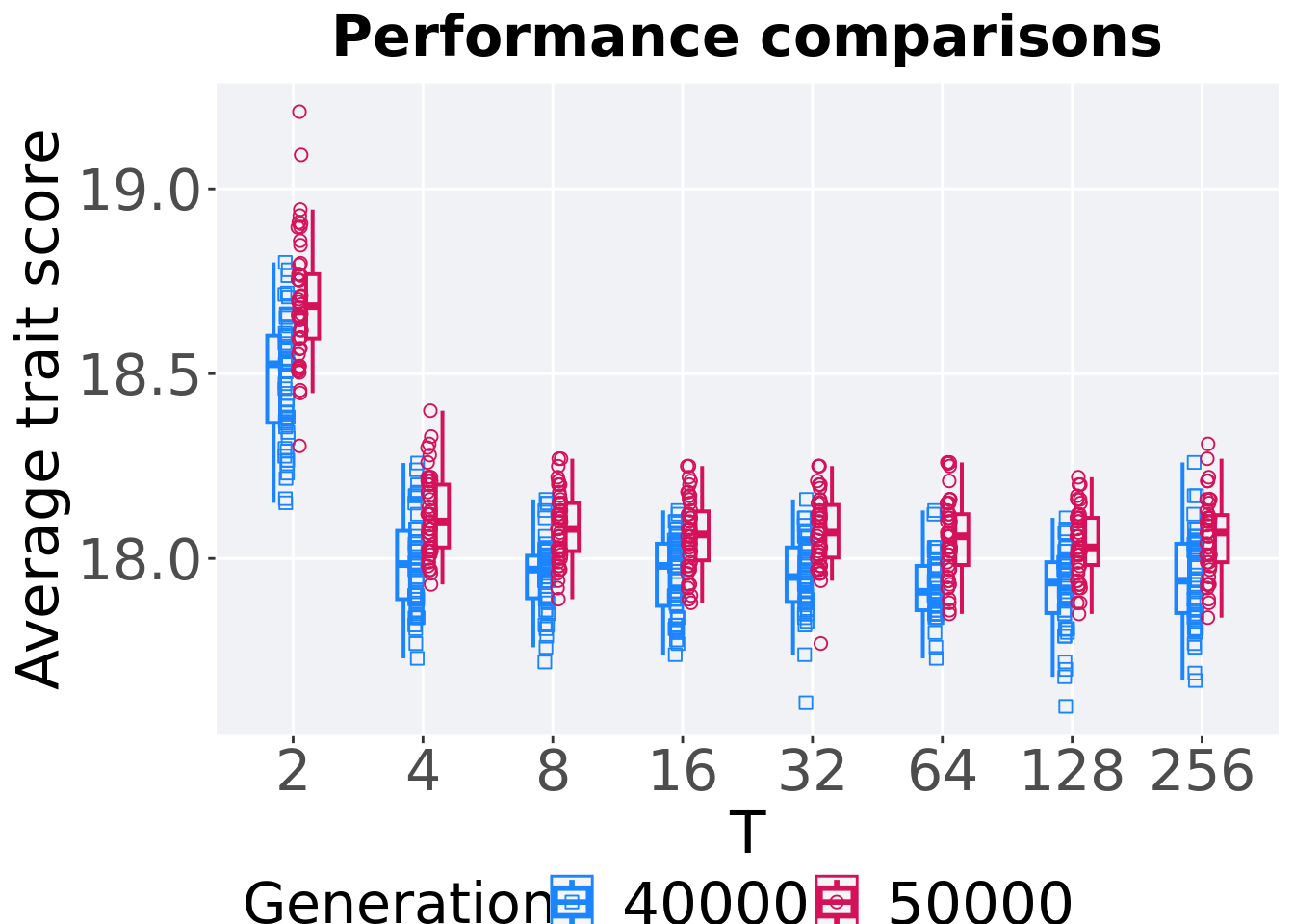
7.1.3.2 Performance comparison
Best performances in the population at 40,000 and 50,000 generations.
# 80% and final generation comparison
end = filter(tor_ot_mvc, diagnostic == 'exploitation_rate' & gen == 50000 & T != 'ran')
end$Generation <- factor(end$gen)
mid = filter(tor_ot_mvc, diagnostic == 'exploitation_rate' & gen == 40000 & T != 'ran')
mid$Generation <- factor(mid$gen)
mvc_p = ggplot(mid, aes(x = T, y=pop_fit_max / DIMENSIONALITY, group = T, shape = Generation)) +
geom_point(col = mvc_col[1] , position = position_jitternudge(jitter.width = .03, nudge.x = -0.05), size = 2, alpha = 1.0) +
geom_boxplot(position = position_nudge(x = -.15, y = 0), lwd = 0.7, col = mvc_col[1], fill = mvc_col[1], width = .1, outlier.shape = NA, alpha = 0.0) +
geom_point(data = end, aes(x = T, y=pop_fit_max / DIMENSIONALITY), col = mvc_col[2], position = position_jitternudge(jitter.width = .03, nudge.x = 0.05), size = 2, alpha = 1.0) +
geom_boxplot(data = end, aes(x = T, y=pop_fit_max / DIMENSIONALITY), position = position_nudge(x = .15, y = 0), lwd = 0.7, col = mvc_col[2], fill = mvc_col[2], width = .1, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_discrete(
name="T"
)+
scale_shape_manual(values=c(0,1))+
scale_colour_manual(values = c(mvc_col[1],mvc_col[2])) +
p_theme
plot_grid(
mvc_p +
ggtitle("Performance comparisons") +
theme(legend.position="none"),
legend,
nrow=2,
rel_heights = c(1,.05),
label_size = TSIZE
)
7.1.3.3 Stats
Summary statistics for the performance of the best performance at 40,000 and 50,000 generations.
slices = filter(tor_ot_mvc, diagnostic == 'exploitation_rate' & (gen == 50000 | gen == 40000))
slices$Generation <- factor(slices$gen, levels = c(50000,40000))
slices %>%
group_by(T, Generation) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_fit_max / DIMENSIONALITY)),
min = min(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
median = median(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
mean = mean(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
max = max(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
IQR = IQR(pop_fit_max / DIMENSIONALITY, na.rm = TRUE)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.## # A tibble: 16 x 9
## # Groups: T [8]
## T Generation count na_cnt min median mean max IQR
## <fct> <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2 50000 50 0 18.3 18.7 18.7 19.2 0.174
## 2 2 40000 50 0 18.2 18.5 18.5 18.8 0.236
## 3 4 50000 50 0 17.9 18.1 18.1 18.4 0.170
## 4 4 40000 50 0 17.7 18.0 18.0 18.3 0.185
## 5 8 50000 50 0 17.9 18.1 18.1 18.3 0.130
## 6 8 40000 50 0 17.7 18.0 18.0 18.2 0.115
## 7 16 50000 50 0 17.9 18.1 18.1 18.2 0.133
## 8 16 40000 50 0 17.7 18.0 18.0 18.1 0.168
## 9 32 50000 50 0 17.8 18.1 18.1 18.2 0.142
## 10 32 40000 50 0 17.6 17.9 18.0 18.2 0.147
## 11 64 50000 50 0 17.8 18.1 18.1 18.3 0.137
## 12 64 40000 50 0 17.7 17.9 17.9 18.1 0.120
## 13 128 50000 50 0 17.8 18.0 18.0 18.2 0.128
## 14 128 40000 50 0 17.6 17.9 17.9 18.1 0.137
## 15 256 50000 50 0 17.8 18.1 18.1 18.3 0.128
## 16 256 40000 50 0 17.7 17.9 18.0 18.3 0.188T 2
wilcox.test(x = filter(slices, T == 2 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 2 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 2 & Generation == 50000)$pop_fit_max and filter(slices, T == 2 & Generation == 40000)$pop_fit_max
## W = 2007, p-value = 1.836e-07
## alternative hypothesis: true location shift is not equal to 0T 4
wilcox.test(x = filter(slices, T == 4 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 4 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 4 & Generation == 50000)$pop_fit_max and filter(slices, T == 4 & Generation == 40000)$pop_fit_max
## W = 1961, p-value = 9.59e-07
## alternative hypothesis: true location shift is not equal to 0T 8
wilcox.test(x = filter(slices, T == 8 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 8 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 8 & Generation == 50000)$pop_fit_max and filter(slices, T == 8 & Generation == 40000)$pop_fit_max
## W = 2075, p-value = 1.301e-08
## alternative hypothesis: true location shift is not equal to 0T 16
wilcox.test(x = filter(slices, T == 16 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 16 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 16 & Generation == 50000)$pop_fit_max and filter(slices, T == 16 & Generation == 40000)$pop_fit_max
## W = 1948.5, p-value = 1.483e-06
## alternative hypothesis: true location shift is not equal to 0T 32
wilcox.test(x = filter(slices, T == 32 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 32 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 32 & Generation == 50000)$pop_fit_max and filter(slices, T == 32 & Generation == 40000)$pop_fit_max
## W = 2020.5, p-value = 1.097e-07
## alternative hypothesis: true location shift is not equal to 0T 64
wilcox.test(x = filter(slices, T == 64 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 64 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 64 & Generation == 50000)$pop_fit_max and filter(slices, T == 64 & Generation == 40000)$pop_fit_max
## W = 2104.5, p-value = 3.84e-09
## alternative hypothesis: true location shift is not equal to 0T 128
wilcox.test(x = filter(slices, T == 128 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 128 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 128 & Generation == 50000)$pop_fit_max and filter(slices, T == 128 & Generation == 40000)$pop_fit_max
## W = 2003.5, p-value = 2.068e-07
## alternative hypothesis: true location shift is not equal to 0T 256
wilcox.test(x = filter(slices, T == 256 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 256 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 256 & Generation == 50000)$pop_fit_max and filter(slices, T == 256 & Generation == 40000)$pop_fit_max
## W = 1892.5, p-value = 9.536e-06
## alternative hypothesis: true location shift is not equal to 07.2 Ordered exploitation results
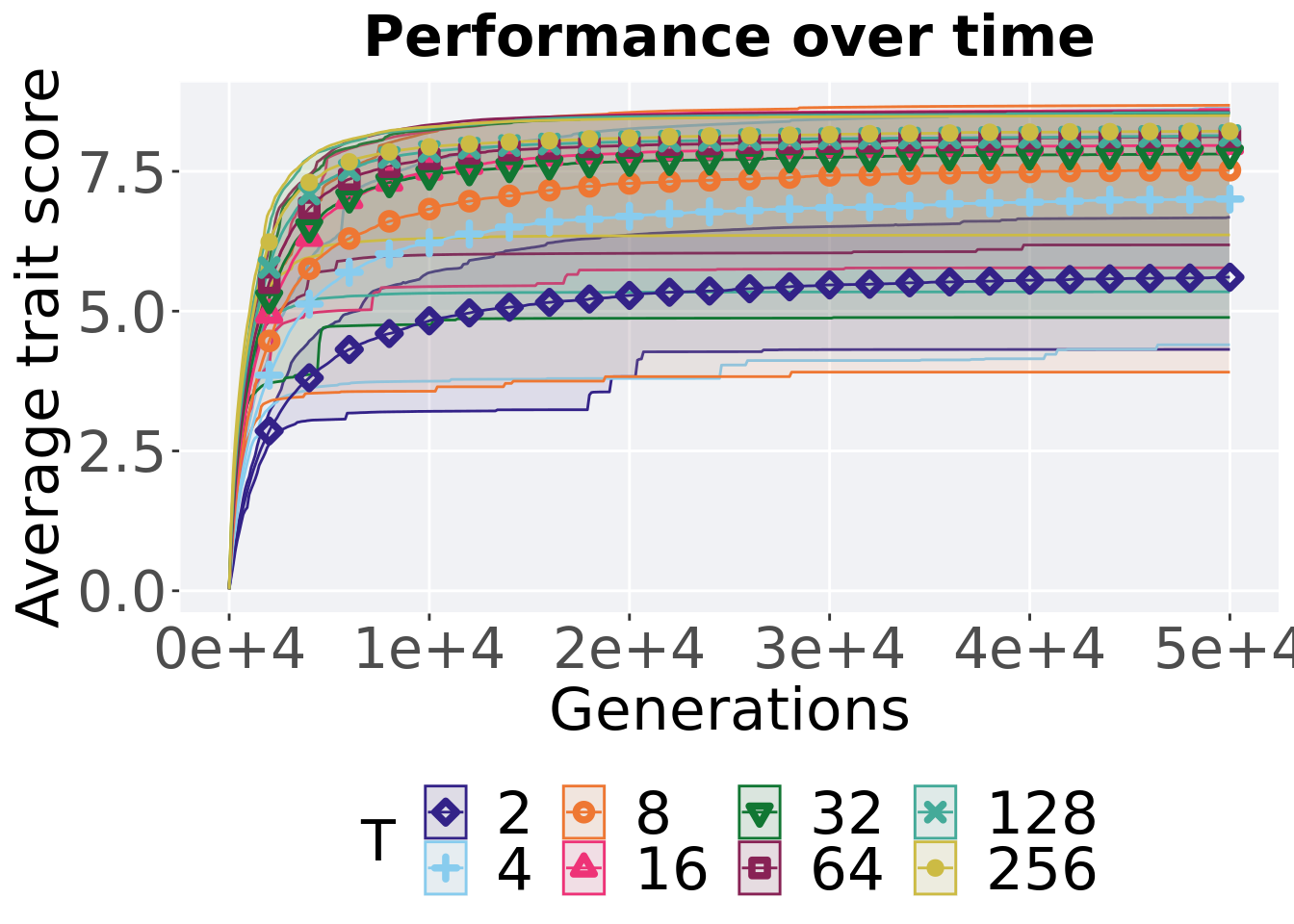
Here we present the results for best performances found by each tournament selection size value replicate on the ordered exploitation diagnostic. Best performance found refers to the largest average trait score found in a given population. Note that performance values fall between 0 and 100.
7.2.1 Performance over time
Performance over time.
lines = filter(tor_ot, diagnostic == 'ordered_exploitation') %>%
group_by(T, gen) %>%
dplyr::summarise(
min = min(pop_fit_max),
mean = mean(pop_fit_max),
max = max(pop_fit_max)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.ggplot(lines, aes(x=gen, y=mean / DIMENSIONALITY, group = T, fill = T, color = T, shape = T)) +
geom_ribbon(aes(ymin = min / DIMENSIONALITY, ymax = max / DIMENSIONALITY), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score",
limits=c(-1, 101),
breaks=seq(0,100, 20),
labels=c("0", "20", "40", "60", "80", "100")
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Best performance over time") +
p_theme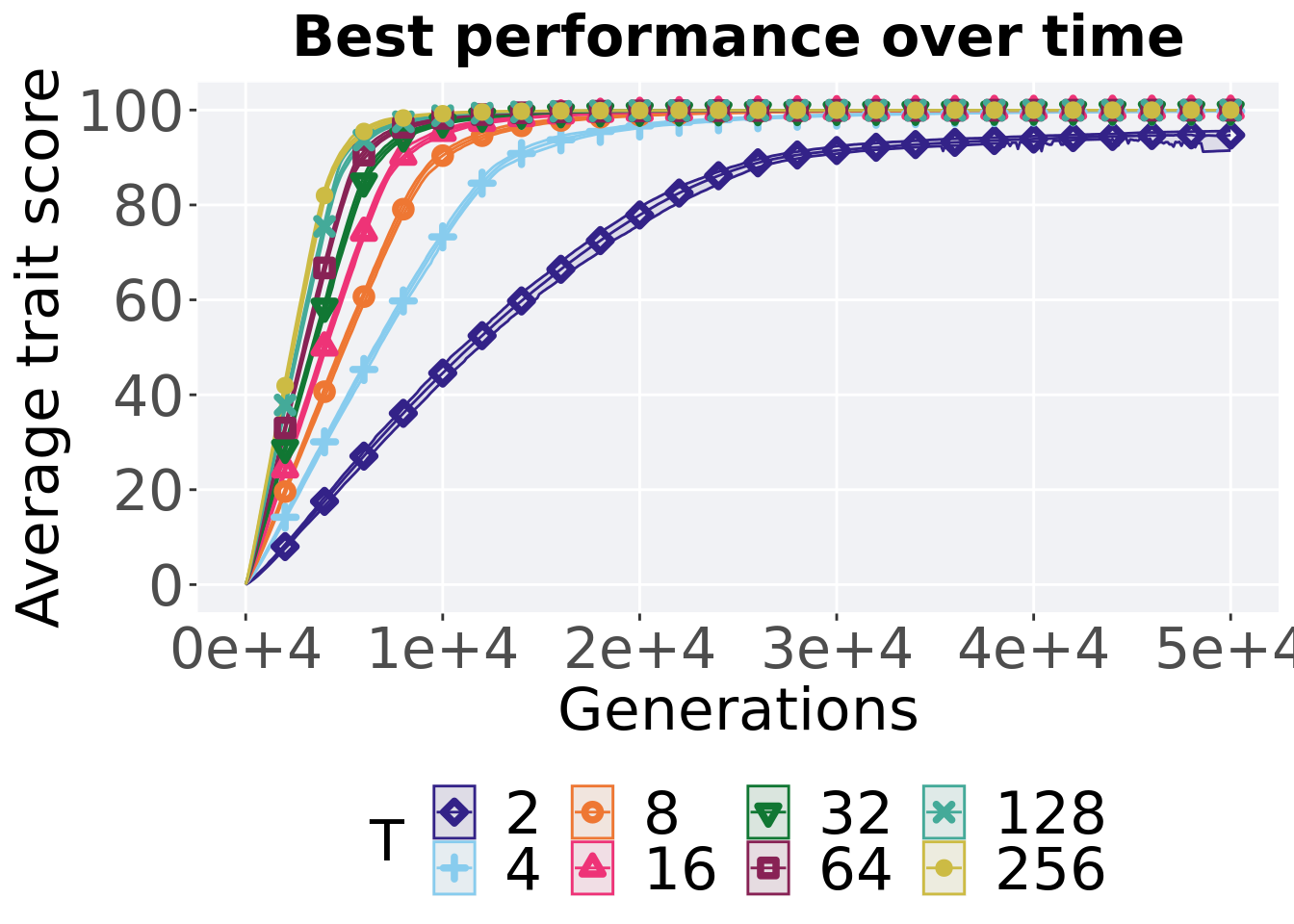
7.2.2 Generation satisfactory solution found
The first Generations a satisfactory solution is found throughout the 50,000 generations.
filter(tor_ssf, Diagnostic == 'ORDERED_EXPLOITATION') %>%
ggplot(., aes(x = T, y = Generations, color = T, fill = T, shape = T)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_shape_manual(values=SHAPE)+
scale_y_continuous(
name="Generation",
limits=c(0, 60000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000, 60000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4", "Fail")
) +
scale_x_discrete(
name="T"
) +
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Generation satisfactory solution found") +
p_theme## Warning: Removed 15 rows containing missing values (`geom_point()`).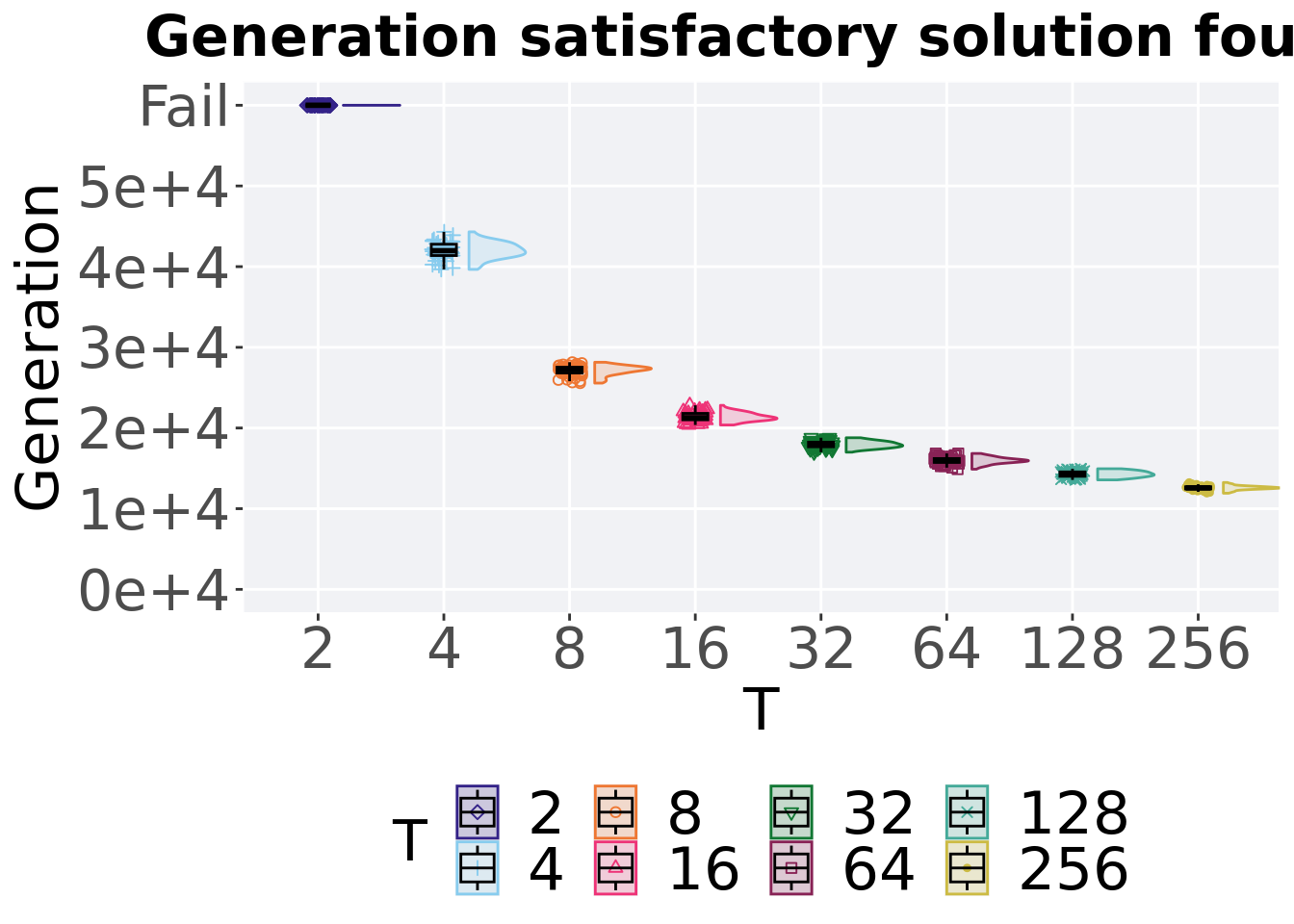
7.2.2.1 Stats
Summary statistics for the first Generations a satisfactory solution is found throughout the 50,000 generations.
ssf = filter(tor_ssf, Diagnostic == 'ORDERED_EXPLOITATION')
group_by(ssf, T) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(Generations)),
min = min(Generations, na.rm = TRUE),
median = median(Generations, na.rm = TRUE),
mean = mean(Generations, na.rm = TRUE),
max = max(Generations, na.rm = TRUE),
IQR = IQR(Generations, na.rm = TRUE)
)## # A tibble: 8 x 8
## T count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 2 50 0 60000 60000 60000 60000 0
## 2 4 50 0 39659 41972. 41991. 44310 1404.
## 3 8 50 0 25563 27254. 27122. 28151 714
## 4 16 50 0 20374 21270. 21366. 22808 792.
## 5 32 50 0 17005 17945 17950. 18792 572.
## 6 64 50 0 14896 15960 15965. 16845 480.
## 7 128 50 0 13569 14278. 14278. 14952 545
## 8 256 50 0 11919 12578. 12580. 13249 262.Kruskal–Wallis test provides evidence of significant differences among the first Generations a satisfactory solution is found throughout the 50,000 generations.
##
## Kruskal-Wallis rank sum test
##
## data: Generations by T
## Kruskal-Wallis chi-squared = 393.52, df = 7, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction on the first Generations a satisfactory solution is found throughout the 50,000 generations.
pairwise.wilcox.test(x = ssf$Generations, g = ssf$T , p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'l')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: ssf$Generations and ssf$T
##
## 2 4 8 16 32 64 128
## 4 <2e-16 - - - - - -
## 8 <2e-16 <2e-16 - - - - -
## 16 <2e-16 <2e-16 <2e-16 - - - -
## 32 <2e-16 <2e-16 <2e-16 <2e-16 - - -
## 64 <2e-16 <2e-16 <2e-16 <2e-16 <2e-16 - -
## 128 <2e-16 <2e-16 <2e-16 <2e-16 <2e-16 <2e-16 -
## 256 <2e-16 <2e-16 <2e-16 <2e-16 <2e-16 <2e-16 <2e-16
##
## P value adjustment method: bonferroni7.2.3 Multi-valley crossing
7.2.3.1 Performance over time
# data for lines and shading on plots
lines = filter(tor_ot_mvc, diagnostic == 'ordered_exploitation') %>%
group_by(T, gen) %>%
dplyr::summarise(
min = min(pop_fit_max) / DIMENSIONALITY,
mean = mean(pop_fit_max) / DIMENSIONALITY,
max = max(pop_fit_max) / DIMENSIONALITY
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.ggplot(lines, aes(x=gen, y=mean, group = T, fill =T, color = T, shape = T)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Performance over time')+
p_theme +
guides(
shape=guide_legend(nrow=2, title.position = "left"),
color=guide_legend(nrow=2, title.position = "left"),
fill=guide_legend(nrow=2, title.position = "left")
)
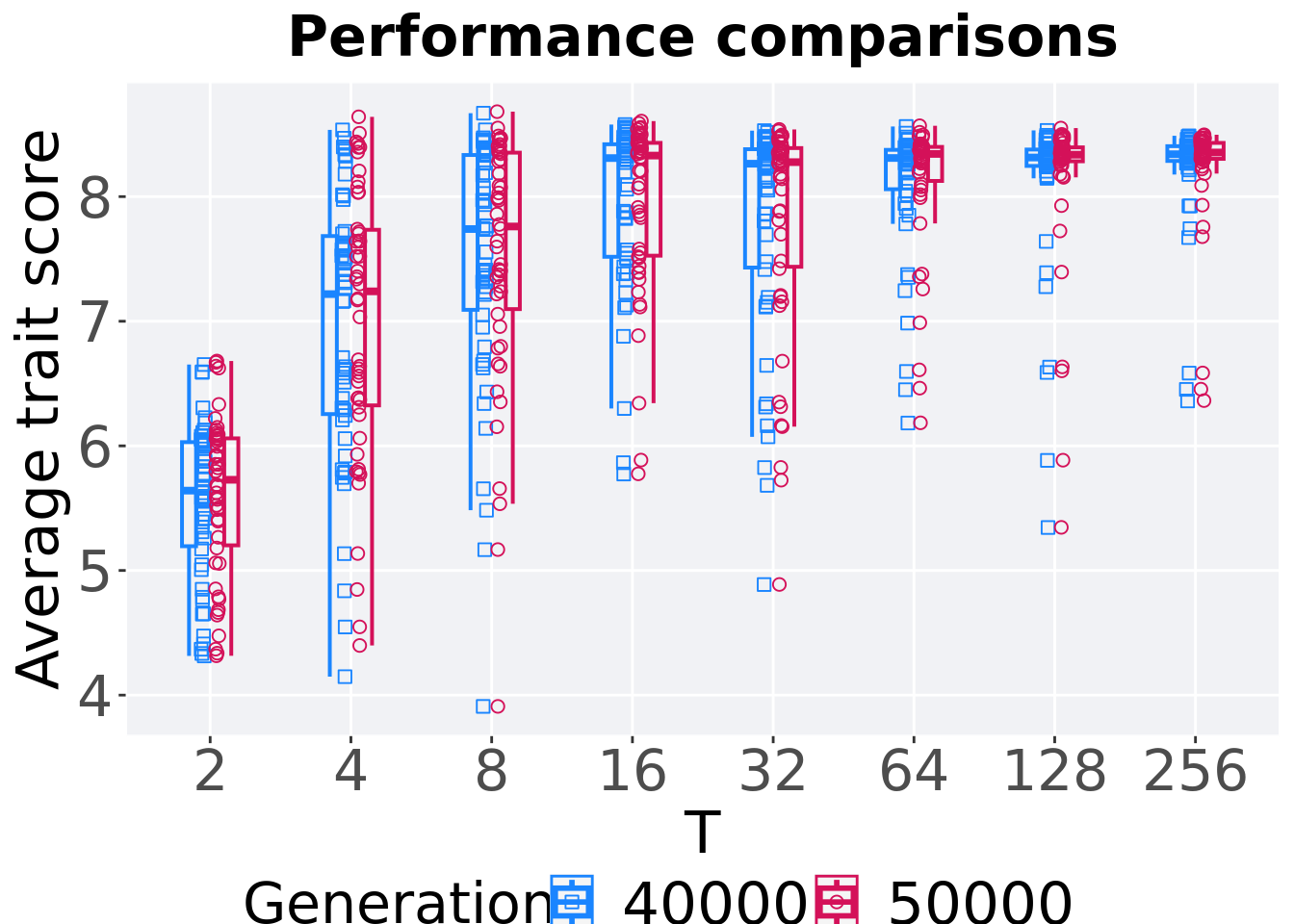
7.2.3.2 Performance comparison
Best performances in the population at 40,000 and 50,000 generations.
# 80% and final generation comparison
end = filter(tor_ot_mvc, diagnostic == 'ordered_exploitation' & gen == 50000 & T != 'ran')
end$Generation <- factor(end$gen)
mid = filter(tor_ot_mvc, diagnostic == 'ordered_exploitation' & gen == 40000 & T != 'ran')
mid$Generation <- factor(mid$gen)
mvc_p = ggplot(mid, aes(x = T, y=pop_fit_max / DIMENSIONALITY, group = T, shape = Generation)) +
geom_point(col = mvc_col[1] , position = position_jitternudge(jitter.width = .03, nudge.x = -0.05), size = 2, alpha = 1.0) +
geom_boxplot(position = position_nudge(x = -.15, y = 0), lwd = 0.7, col = mvc_col[1], fill = mvc_col[1], width = .1, outlier.shape = NA, alpha = 0.0) +
geom_point(data = end, aes(x = T, y=pop_fit_max / DIMENSIONALITY), col = mvc_col[2], position = position_jitternudge(jitter.width = .03, nudge.x = 0.05), size = 2, alpha = 1.0) +
geom_boxplot(data = end, aes(x = T, y=pop_fit_max / DIMENSIONALITY), position = position_nudge(x = .15, y = 0), lwd = 0.7, col = mvc_col[2], fill = mvc_col[2], width = .1, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_discrete(
name="T"
)+
scale_shape_manual(values=c(0,1))+
scale_colour_manual(values = c(mvc_col[1],mvc_col[2])) +
p_theme
plot_grid(
mvc_p +
ggtitle("Performance comparisons") +
theme(legend.position="none"),
legend,
nrow=2,
rel_heights = c(1,.05),
label_size = TSIZE
)
7.2.3.3 Stats
Summary statistics for the performance of the best performance at 40,000 and 50,000 generations.
slices = filter(tor_ot_mvc, diagnostic == 'ordered_exploitation' & (gen == 50000 | gen == 40000))
slices$Generation <- factor(slices$gen, levels = c(50000,40000))
slices %>%
group_by(T, Generation) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_fit_max / DIMENSIONALITY)),
min = min(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
median = median(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
mean = mean(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
max = max(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
IQR = IQR(pop_fit_max / DIMENSIONALITY, na.rm = TRUE)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.## # A tibble: 16 x 9
## # Groups: T [8]
## T Generation count na_cnt min median mean max IQR
## <fct> <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2 50000 50 0 4.32 5.73 5.61 6.68 0.858
## 2 2 40000 50 0 4.31 5.64 5.55 6.65 0.835
## 3 4 50000 50 0 4.40 7.24 7.00 8.64 1.41
## 4 4 40000 50 0 4.15 7.22 6.95 8.54 1.43
## 5 8 50000 50 0 3.91 7.76 7.52 8.68 1.26
## 6 8 40000 50 0 3.91 7.74 7.49 8.67 1.24
## 7 16 50000 50 0 5.78 8.33 7.96 8.61 0.905
## 8 16 40000 50 0 5.77 8.31 7.95 8.58 0.903
## 9 32 50000 50 0 4.89 8.28 7.81 8.54 0.952
## 10 32 40000 50 0 4.89 8.26 7.79 8.53 0.950
## 11 64 50000 50 0 6.18 8.34 8.12 8.57 0.273
## 12 64 40000 50 0 6.18 8.31 8.09 8.56 0.316
## 13 128 50000 50 0 5.35 8.34 8.14 8.55 0.112
## 14 128 40000 50 0 5.34 8.31 8.11 8.53 0.132
## 15 256 50000 50 0 6.36 8.35 8.22 8.50 0.128
## 16 256 40000 50 0 6.36 8.34 8.20 8.49 0.119T 2
wilcox.test(x = filter(slices, T == 2 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 2 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 2 & Generation == 50000)$pop_fit_max and filter(slices, T == 2 & Generation == 40000)$pop_fit_max
## W = 1337, p-value = 0.551
## alternative hypothesis: true location shift is not equal to 0T 4
wilcox.test(x = filter(slices, T == 4 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 4 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 4 & Generation == 50000)$pop_fit_max and filter(slices, T == 4 & Generation == 40000)$pop_fit_max
## W = 1315, p-value = 0.6566
## alternative hypothesis: true location shift is not equal to 0T 8
wilcox.test(x = filter(slices, T == 8 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 8 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 8 & Generation == 50000)$pop_fit_max and filter(slices, T == 8 & Generation == 40000)$pop_fit_max
## W = 1306.5, p-value = 0.6995
## alternative hypothesis: true location shift is not equal to 0T 16
wilcox.test(x = filter(slices, T == 16 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 16 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 16 & Generation == 50000)$pop_fit_max and filter(slices, T == 16 & Generation == 40000)$pop_fit_max
## W = 1309, p-value = 0.6867
## alternative hypothesis: true location shift is not equal to 0T 32
wilcox.test(x = filter(slices, T == 32 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 32 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 32 & Generation == 50000)$pop_fit_max and filter(slices, T == 32 & Generation == 40000)$pop_fit_max
## W = 1305, p-value = 0.7071
## alternative hypothesis: true location shift is not equal to 0T 64
wilcox.test(x = filter(slices, T == 64 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 64 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 64 & Generation == 50000)$pop_fit_max and filter(slices, T == 64 & Generation == 40000)$pop_fit_max
## W = 1379, p-value = 0.3757
## alternative hypothesis: true location shift is not equal to 0T 128
wilcox.test(x = filter(slices, T == 128 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 128 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 128 & Generation == 50000)$pop_fit_max and filter(slices, T == 128 & Generation == 40000)$pop_fit_max
## W = 1409, p-value = 0.2745
## alternative hypothesis: true location shift is not equal to 0T 256
wilcox.test(x = filter(slices, T == 256 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 256 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 256 & Generation == 50000)$pop_fit_max and filter(slices, T == 256 & Generation == 40000)$pop_fit_max
## W = 1381, p-value = 0.3683
## alternative hypothesis: true location shift is not equal to 07.3 Contraditory objectives diagnostic
Here we present the results for satisfactory trait coverage and activation gene coverage found by each tournament selection size value replicate on the ordered exploitation diagnostic. Satisfactory trait coverage refers to the count of unique satisfied traits in the population, while activation gene coverage refers to the count of unique activation genes in the population. Note that both coverage values fall between 0 and 100.
7.3.1 Satisfactory trait coverage
Satisfactory trait coverage analysis.
7.3.1.1 Coverage over time
Satisfactory trait coverage over time.
lines = filter(tor_ot, diagnostic == 'contradictory_objectives') %>%
group_by(T, gen) %>%
dplyr::summarise(
min = min(pop_uni_obj),
mean = mean(pop_uni_obj),
max = max(pop_uni_obj)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.ggplot(lines, aes(x=gen, y=mean, group = T, fill =T, color = T, shape = T)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Satisfactory trait coverage over time") +
p_theme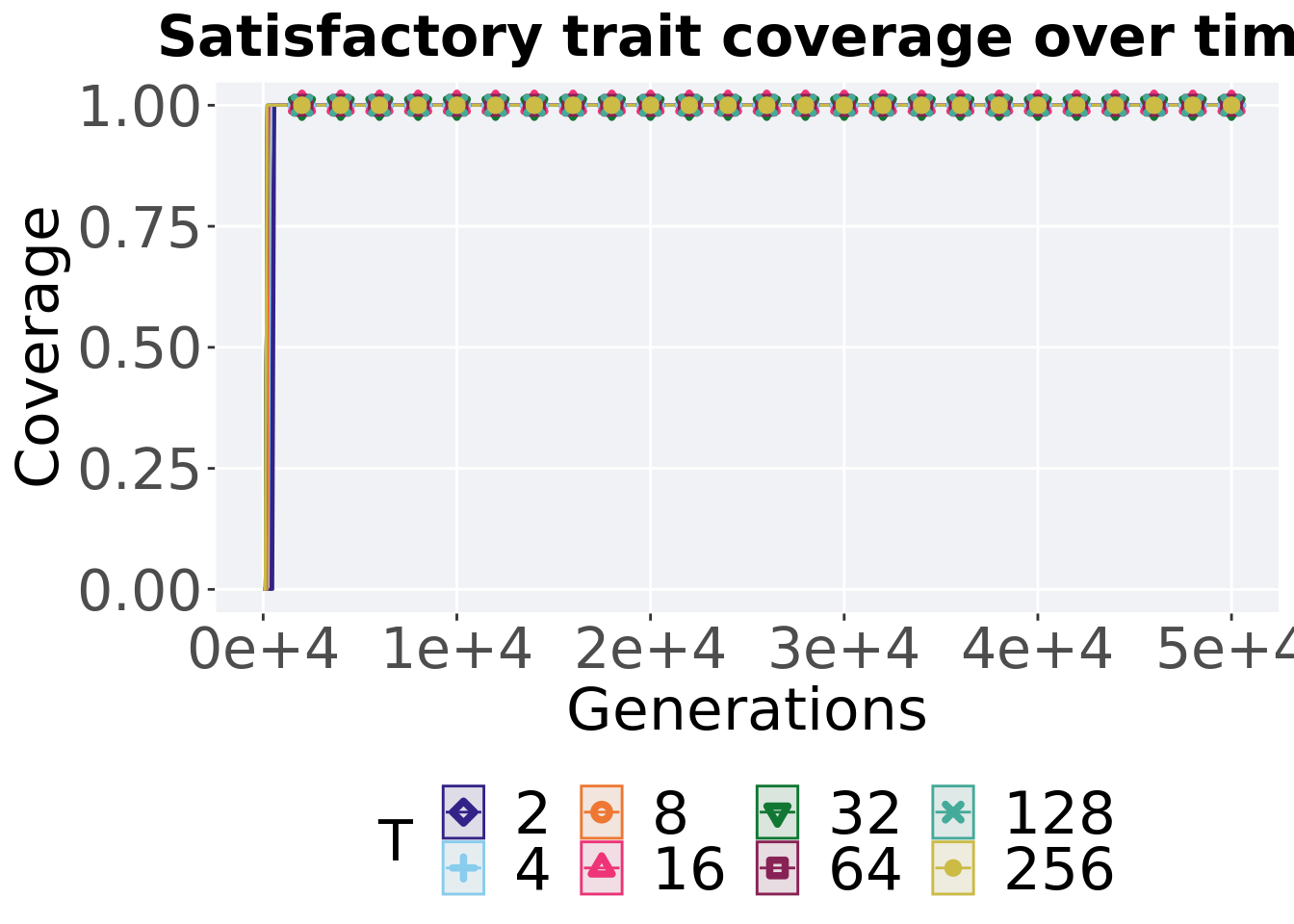
7.3.1.2 Best coverage throughout
Best satisfactory trait coverage throughout 50,000 generations.
filter(tor_best, col == 'pop_uni_obj' & diagnostic == 'contradictory_objectives') %>%
ggplot(., aes(x = T, y = val, color = T, fill = T, shape = T)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Coverage"
) +
scale_x_discrete(
name="T"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Best satisfactory trait coverage") +
p_theme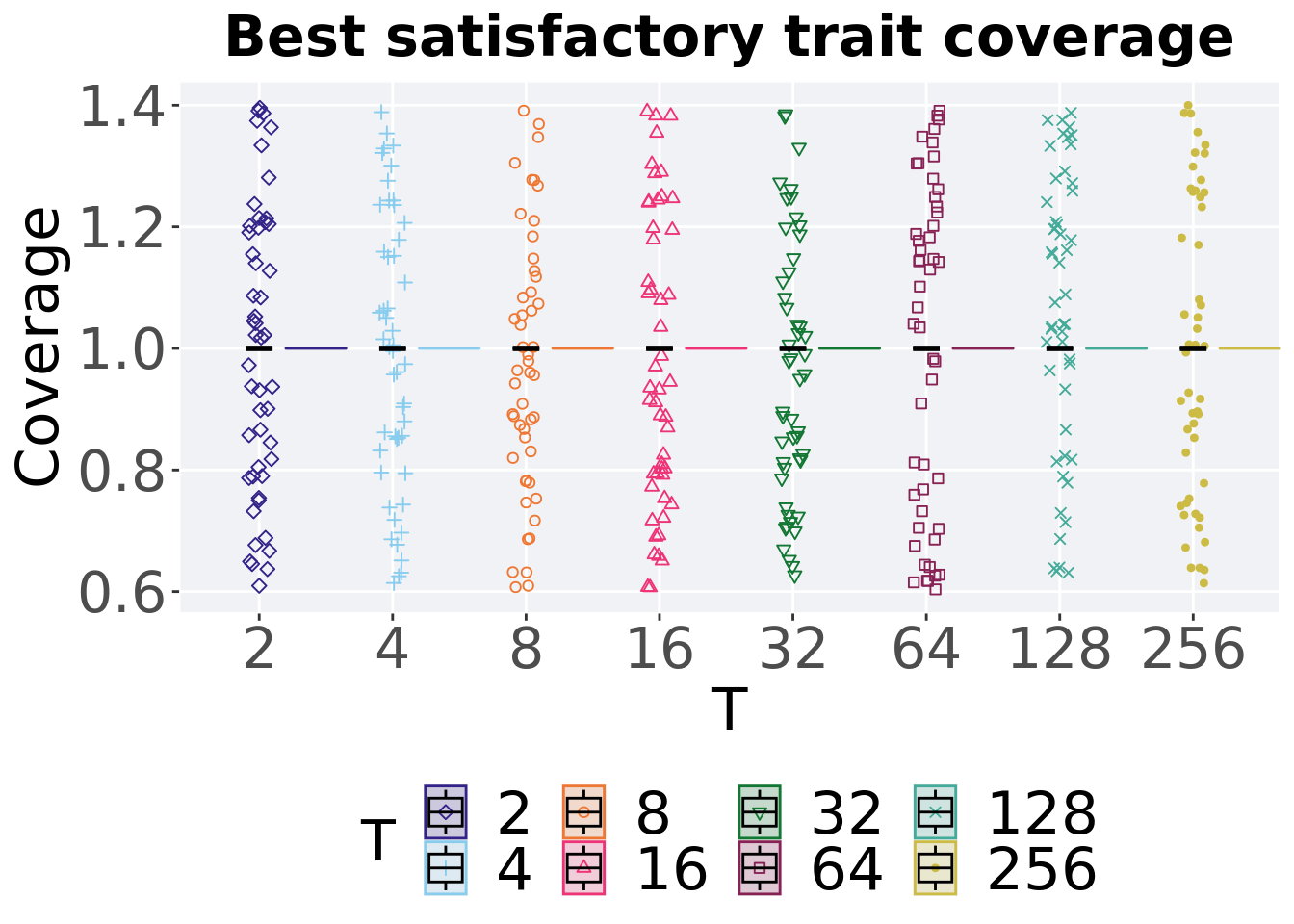
7.3.1.2.1 Stats
Summary statistics for the best satisfactory trait coverage throughout 50,000 generations.
coverage = filter(tor_best, col == 'pop_uni_obj' & diagnostic == 'contradictory_objectives')
group_by(coverage, T) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(val)),
min = min(val, na.rm = TRUE),
median = median(val, na.rm = TRUE),
mean = mean(val, na.rm = TRUE),
max = max(val, na.rm = TRUE),
IQR = IQR(val, na.rm = TRUE)
)## # A tibble: 8 x 8
## T count na_cnt min median mean max IQR
## <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2 50 0 1 1 1 1 0
## 2 4 50 0 1 1 1 1 0
## 3 8 50 0 1 1 1 1 0
## 4 16 50 0 1 1 1 1 0
## 5 32 50 0 1 1 1 1 0
## 6 64 50 0 1 1 1 1 0
## 7 128 50 0 1 1 1 1 0
## 8 256 50 0 1 1 1 1 07.3.1.3 End of 50,000 generations
Satisfactory trait coverage in the population at the end of 50,000 generations.
filter(tor_ot, diagnostic == 'contradictory_objectives' & gen == 50000) %>%
ggplot(., aes(x = T, y = pop_uni_obj, color = T, fill = T, shape = T)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Coverage"
) +
scale_x_discrete(
name="T"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Final satisfactory trait coverage") +
p_theme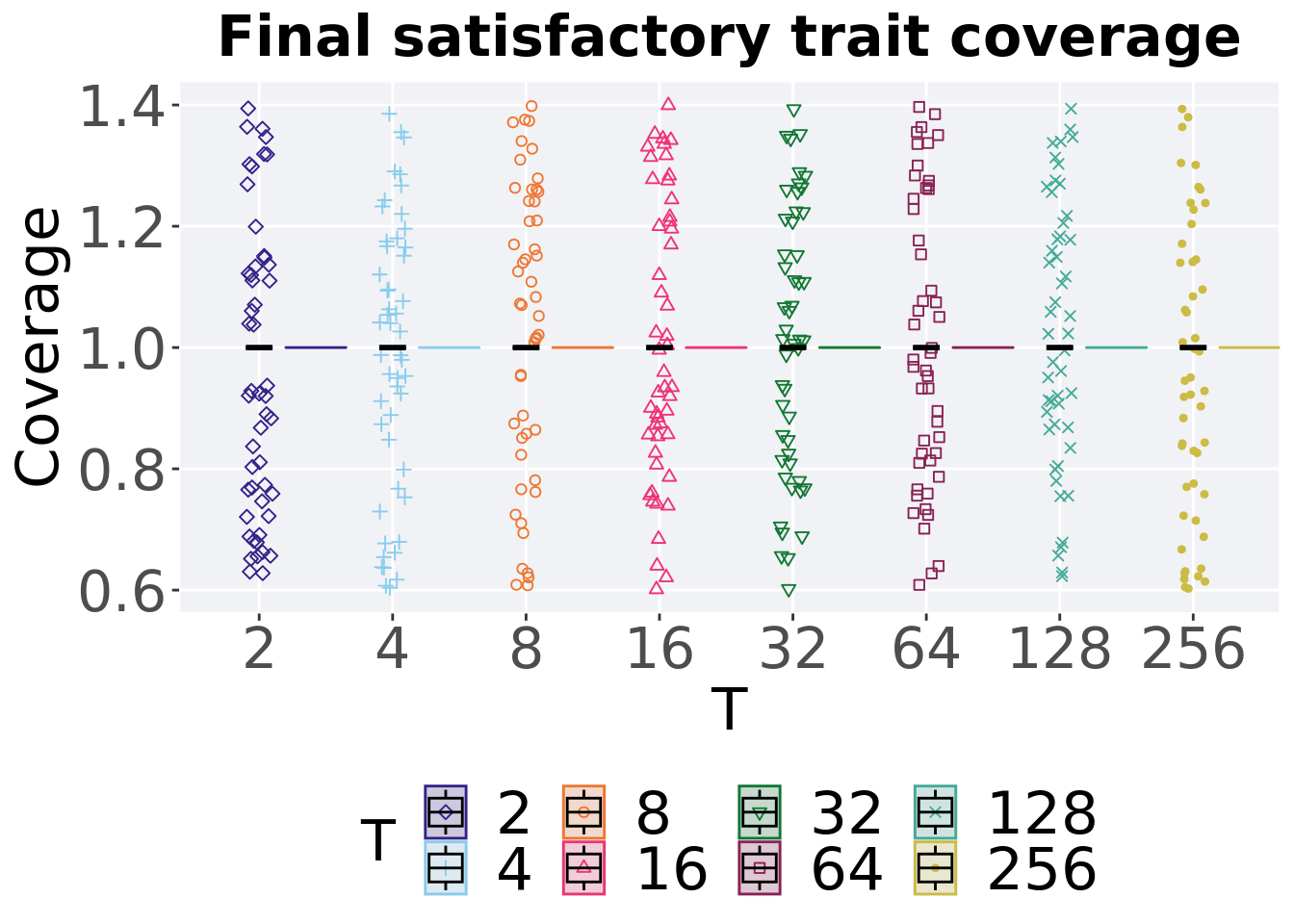
7.3.1.3.1 Stats
Summary statistics for satisfactory trait coverage in the population at the end of 50,000 generations.
coverage = filter(tor_ot, diagnostic == 'contradictory_objectives' & gen == 50000)
group_by(coverage, T) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_uni_obj)),
min = min(pop_uni_obj, na.rm = TRUE),
median = median(pop_uni_obj, na.rm = TRUE),
mean = mean(pop_uni_obj, na.rm = TRUE),
max = max(pop_uni_obj, na.rm = TRUE),
IQR = IQR(pop_uni_obj, na.rm = TRUE)
)## # A tibble: 8 x 8
## T count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 2 50 0 1 1 1 1 0
## 2 4 50 0 1 1 1 1 0
## 3 8 50 0 1 1 1 1 0
## 4 16 50 0 1 1 1 1 0
## 5 32 50 0 1 1 1 1 0
## 6 64 50 0 1 1 1 1 0
## 7 128 50 0 1 1 1 1 0
## 8 256 50 0 1 1 1 1 07.3.2 Activation gene coverage
Here we analyze the activation gene coverage for each parameter replicate on the contradictory objectives diagnostic.
7.3.2.1 Coverage over time
Activation gene coverage over time.
lines = filter(tor_ot, diagnostic == 'contradictory_objectives') %>%
group_by(T, gen) %>%
dplyr::summarise(
min = min(uni_str_pos),
mean = mean(uni_str_pos),
max = max(uni_str_pos)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.ggplot(lines, aes(x=gen, y=mean, group = T, fill =T, color = T, shape = T)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage",
limits=c(-1, 101),
breaks=seq(0,100, 20),
labels=c("0", "20", "40", "60", "80", "100")
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Activation gene coverage over time") +
p_theme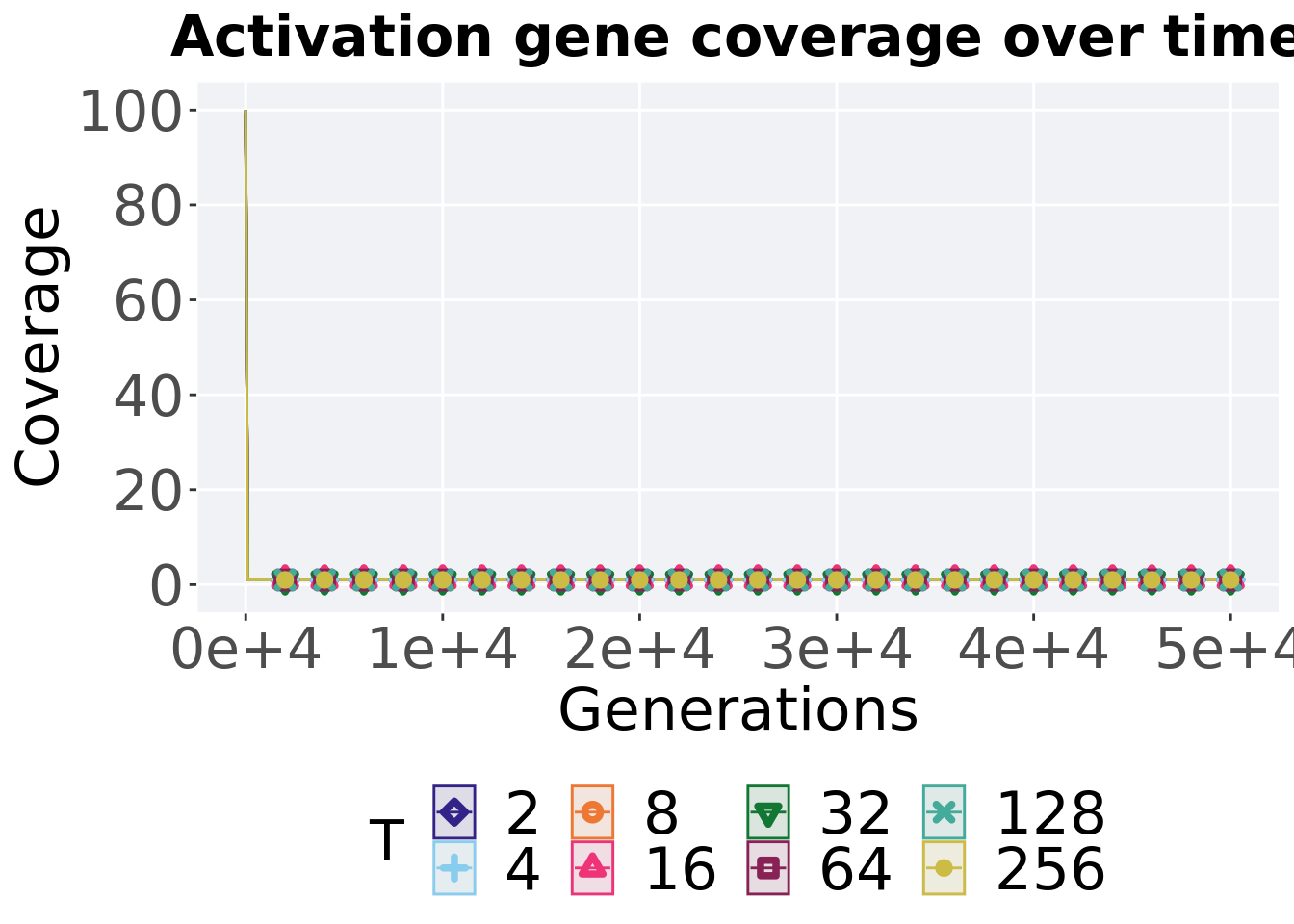
7.3.2.2 End of 50,000 generations
Activation gene coverage in the population at the end of 50,000 generations.
filter(tor_ot, diagnostic == 'contradictory_objectives' & gen == 50000) %>%
ggplot(., aes(x = T, y = uni_str_pos, color = T, fill = T, shape = T)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Coverage"
) +
scale_x_discrete(
name="T"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Final activation gene coverage") +
p_theme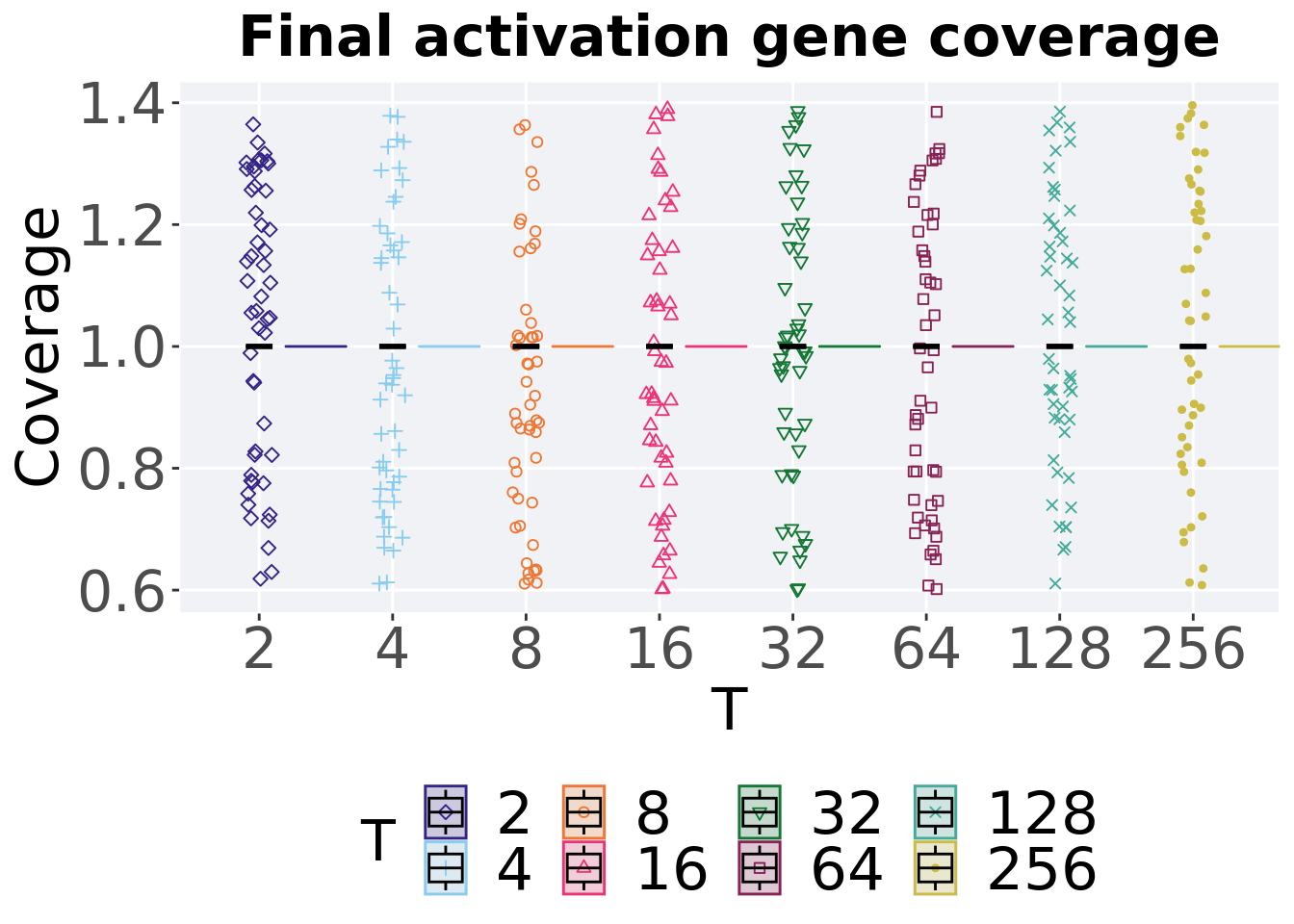
7.3.2.2.1 Stats
Summary statistics for activation gene coverage in the population at the end of 50,000 generations.
coverage = filter(tor_ot, diagnostic == 'contradictory_objectives' & gen == 50000)
group_by(coverage, T) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(uni_str_pos)),
min = min(uni_str_pos, na.rm = TRUE),
median = median(uni_str_pos, na.rm = TRUE),
mean = mean(uni_str_pos, na.rm = TRUE),
max = max(uni_str_pos, na.rm = TRUE),
IQR = IQR(uni_str_pos, na.rm = TRUE)
)## # A tibble: 8 x 8
## T count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 2 50 0 1 1 1 1 0
## 2 4 50 0 1 1 1 1 0
## 3 8 50 0 1 1 1 1 0
## 4 16 50 0 1 1 1 1 0
## 5 32 50 0 1 1 1 1 0
## 6 64 50 0 1 1 1 1 0
## 7 128 50 0 1 1 1 1 0
## 8 256 50 0 1 1 1 1 07.3.3 Multi-valley crossing
7.3.3.1 Satisfactory trait coverage over time
lines = filter(tor_ot_mvc, diagnostic == 'contradictory_objectives') %>%
group_by(T, gen) %>%
dplyr::summarise(
min = min(pop_uni_obj),
mean = mean(pop_uni_obj),
max = max(pop_uni_obj)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.ggplot(lines, aes(x=gen, y=mean, group = T, fill =T, color = T, shape = T)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Satisfactory trait coverage over time") +
p_theme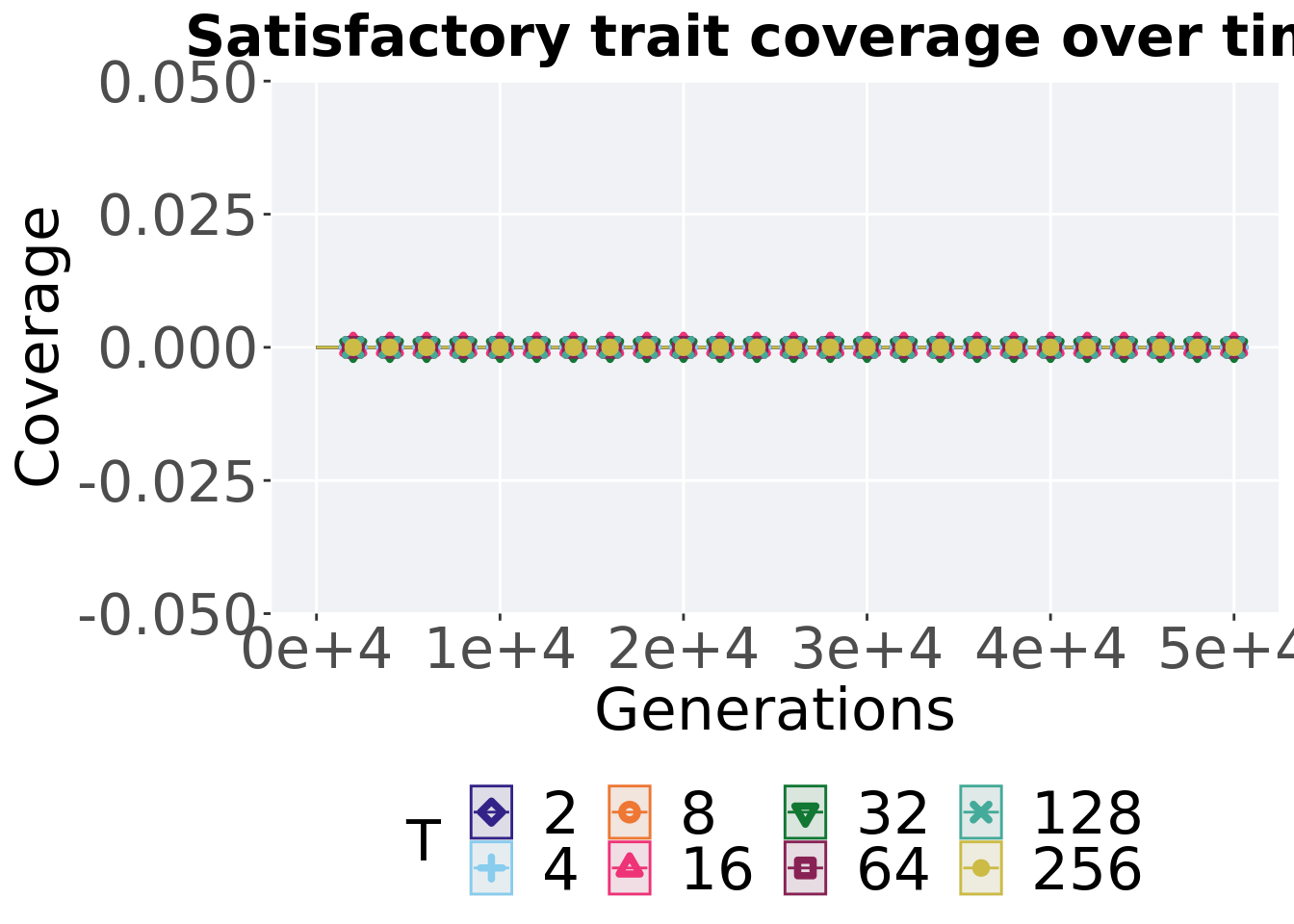
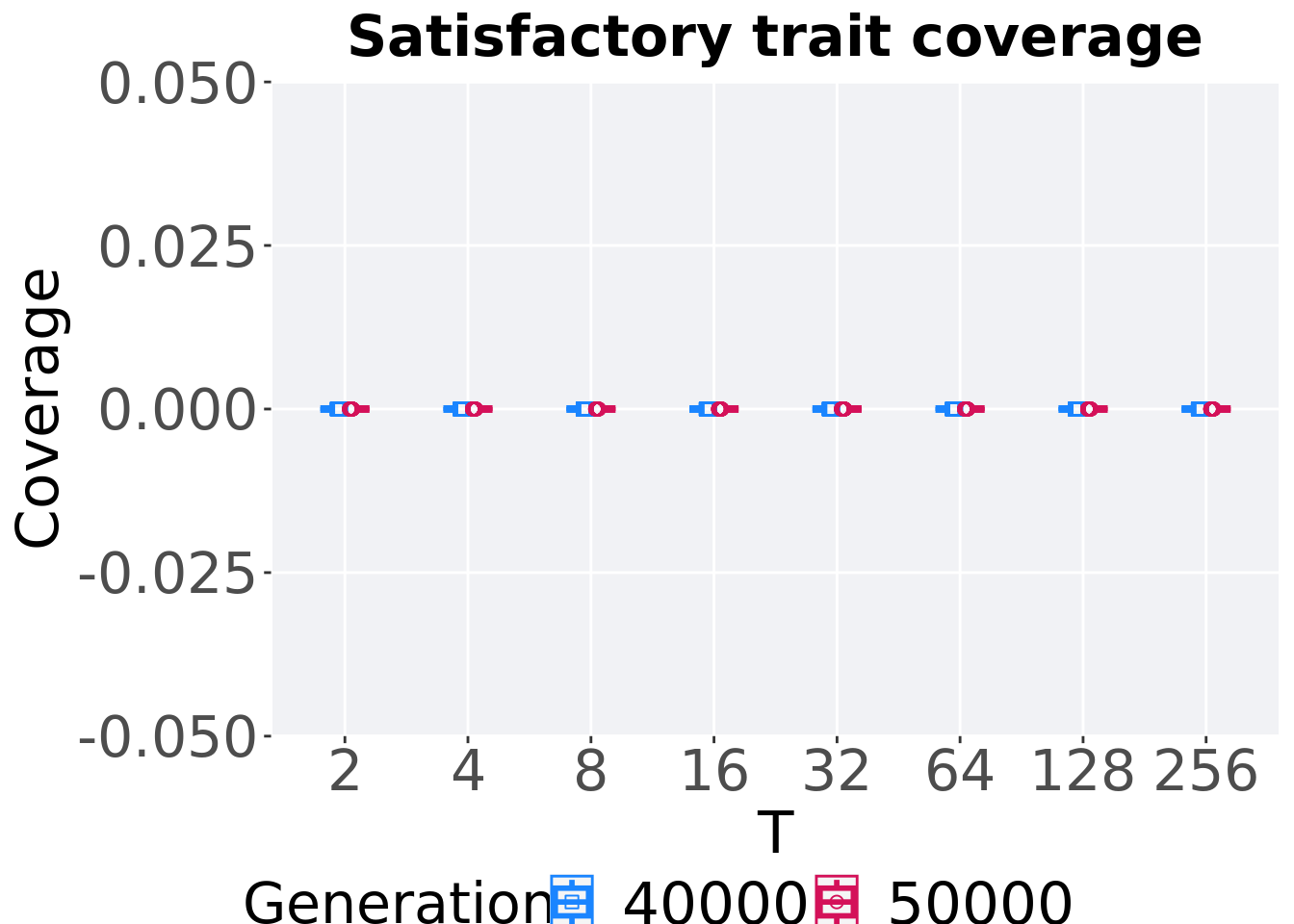
7.3.3.2 Satisfactory trait coverage comparison
Best performances in the population at 40,000 and 50,000 generations.
# 80% and final generation comparison
end = filter(tor_ot_mvc, diagnostic == 'contradictory_objectives' & gen == 50000)
end$Generation <- factor(end$gen)
mid = filter(tor_ot_mvc, diagnostic == 'contradictory_objectives' & gen == 40000)
mid$Generation <- factor(mid$gen)
mvc_p = ggplot(mid, aes(x = T, y=pop_uni_obj, group = T, shape = Generation)) +
geom_point(col = mvc_col[1] , position = position_jitternudge(jitter.width = .03, nudge.x = -0.05), size = 2, alpha = 1.0) +
geom_boxplot(position = position_nudge(x = -.15, y = 0), lwd = 0.7, col = mvc_col[1], fill = mvc_col[1], width = .1, outlier.shape = NA, alpha = 0.0) +
geom_point(data = end, aes(x = T, y=pop_uni_obj), col = mvc_col[2], position = position_jitternudge(jitter.width = .03, nudge.x = 0.05), size = 2, alpha = 1.0) +
geom_boxplot(data = end, aes(x = T, y=pop_uni_obj), position = position_nudge(x = .15, y = 0), lwd = 0.7, col = mvc_col[2], fill = mvc_col[2], width = .1, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Coverage",
) +
scale_x_discrete(
name="T"
)+
scale_shape_manual(values=c(0,1))+
scale_colour_manual(values = c(mvc_col[1],mvc_col[2])) +
p_theme
plot_grid(
mvc_p +
ggtitle("Satisfactory trait coverage") +
theme(legend.position="none"),
legend,
nrow=2,
rel_heights = c(1,.05),
label_size = TSIZE
)
7.3.3.2.1 Stats
Summary statistics for the performance of the best performance at 40,000 and 50,000 generations.
slices = filter(tor_ot_mvc, diagnostic == 'contradictory_objectives' & (gen == 50000 | gen == 40000))
slices$Generation <- factor(slices$gen, levels = c(50000,40000))
slices %>%
group_by(T, Generation) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_uni_obj)),
min = min(pop_uni_obj, na.rm = TRUE),
median = median(pop_uni_obj, na.rm = TRUE),
mean = mean(pop_uni_obj, na.rm = TRUE),
max = max(pop_uni_obj, na.rm = TRUE),
IQR = IQR(pop_uni_obj, na.rm = TRUE)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.## # A tibble: 16 x 9
## # Groups: T [8]
## T Generation count na_cnt min median mean max IQR
## <fct> <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 2 50000 50 0 0 0 0 0 0
## 2 2 40000 50 0 0 0 0 0 0
## 3 4 50000 50 0 0 0 0 0 0
## 4 4 40000 50 0 0 0 0 0 0
## 5 8 50000 50 0 0 0 0 0 0
## 6 8 40000 50 0 0 0 0 0 0
## 7 16 50000 50 0 0 0 0 0 0
## 8 16 40000 50 0 0 0 0 0 0
## 9 32 50000 50 0 0 0 0 0 0
## 10 32 40000 50 0 0 0 0 0 0
## 11 64 50000 50 0 0 0 0 0 0
## 12 64 40000 50 0 0 0 0 0 0
## 13 128 50000 50 0 0 0 0 0 0
## 14 128 40000 50 0 0 0 0 0 0
## 15 256 50000 50 0 0 0 0 0 0
## 16 256 40000 50 0 0 0 0 0 0T 2
wilcox.test(x = filter(slices, T == 2 & Generation == 50000)$pop_uni_obj,
y = filter(slices, T == 2 & Generation == 40000)$pop_uni_obj,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 2 & Generation == 50000)$pop_uni_obj and filter(slices, T == 2 & Generation == 40000)$pop_uni_obj
## W = 1250, p-value = NA
## alternative hypothesis: true location shift is not equal to 0T 4
wilcox.test(x = filter(slices, T == 4 & Generation == 50000)$pop_uni_obj,
y = filter(slices, T == 4 & Generation == 40000)$pop_uni_obj,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 4 & Generation == 50000)$pop_uni_obj and filter(slices, T == 4 & Generation == 40000)$pop_uni_obj
## W = 1250, p-value = NA
## alternative hypothesis: true location shift is not equal to 0T 8
wilcox.test(x = filter(slices, T == 8 & Generation == 50000)$pop_uni_obj,
y = filter(slices, T == 8 & Generation == 40000)$pop_uni_obj,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 8 & Generation == 50000)$pop_uni_obj and filter(slices, T == 8 & Generation == 40000)$pop_uni_obj
## W = 1250, p-value = NA
## alternative hypothesis: true location shift is not equal to 0T 16
wilcox.test(x = filter(slices, T == 16 & Generation == 50000)$pop_uni_obj,
y = filter(slices, T == 16 & Generation == 40000)$pop_uni_obj,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 16 & Generation == 50000)$pop_uni_obj and filter(slices, T == 16 & Generation == 40000)$pop_uni_obj
## W = 1250, p-value = NA
## alternative hypothesis: true location shift is not equal to 0T 32
wilcox.test(x = filter(slices, T == 32 & Generation == 50000)$pop_uni_obj,
y = filter(slices, T == 32 & Generation == 40000)$pop_uni_obj,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 32 & Generation == 50000)$pop_uni_obj and filter(slices, T == 32 & Generation == 40000)$pop_uni_obj
## W = 1250, p-value = NA
## alternative hypothesis: true location shift is not equal to 0T 64
wilcox.test(x = filter(slices, T == 64 & Generation == 50000)$pop_uni_obj,
y = filter(slices, T == 64 & Generation == 40000)$pop_uni_obj,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 64 & Generation == 50000)$pop_uni_obj and filter(slices, T == 64 & Generation == 40000)$pop_uni_obj
## W = 1250, p-value = NA
## alternative hypothesis: true location shift is not equal to 0T 128
wilcox.test(x = filter(slices, T == 128 & Generation == 50000)$pop_uni_obj,
y = filter(slices, T == 128 & Generation == 40000)$pop_uni_obj,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 128 & Generation == 50000)$pop_uni_obj and filter(slices, T == 128 & Generation == 40000)$pop_uni_obj
## W = 1250, p-value = NA
## alternative hypothesis: true location shift is not equal to 0T 256
wilcox.test(x = filter(slices, T == 256 & Generation == 50000)$pop_uni_obj,
y = filter(slices, T == 256 & Generation == 40000)$pop_uni_obj,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 256 & Generation == 50000)$pop_uni_obj and filter(slices, T == 256 & Generation == 40000)$pop_uni_obj
## W = 1250, p-value = NA
## alternative hypothesis: true location shift is not equal to 07.3.3.3 Activation gene coverage over time
lines = filter(tor_ot_mvc, diagnostic == 'contradictory_objectives') %>%
group_by(T, gen) %>%
dplyr::summarise(
min = min(uni_str_pos),
mean = mean(uni_str_pos),
max = max(uni_str_pos)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.ggplot(lines, aes(x=gen, y=mean, group = T, fill =T, color = T, shape = T)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Activation gene coverage over time") +
p_theme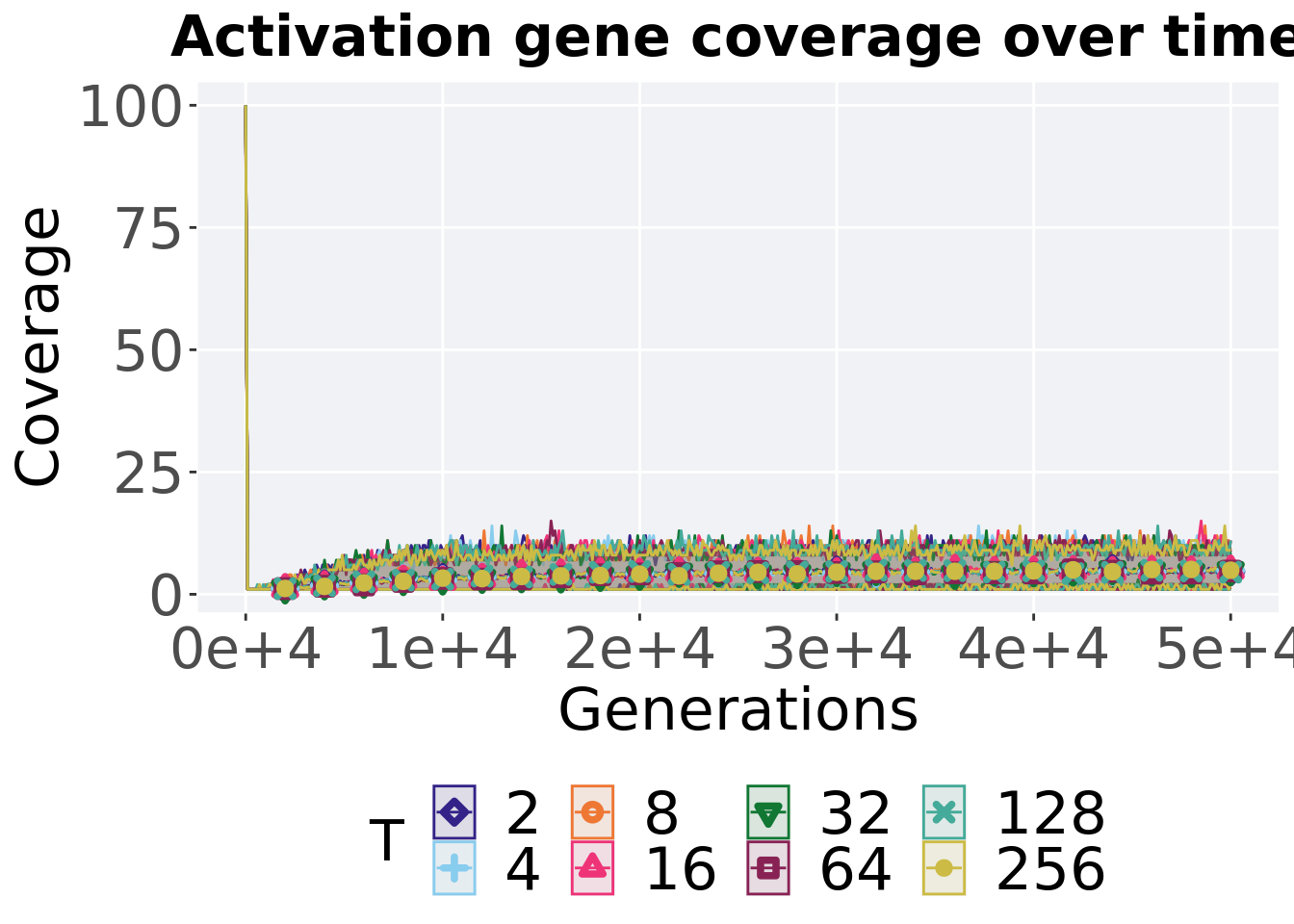
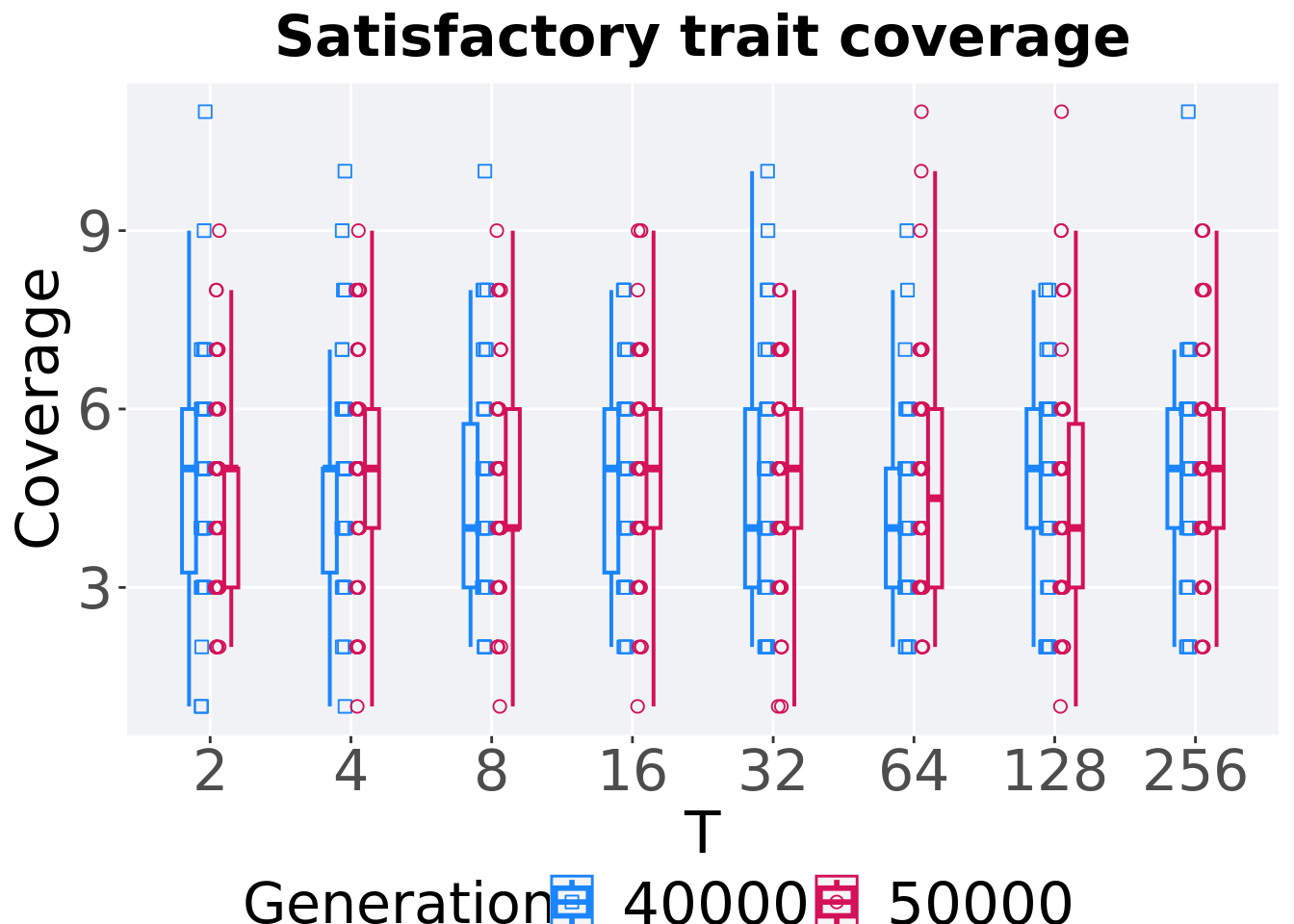
7.3.3.4 Activation gene coverage comparison
Activation gene coverage in the population at 40,000 and 50,000 generations.
# 80% and final generation comparison
end = filter(tor_ot_mvc, diagnostic == 'contradictory_objectives' & gen == 50000)
end$Generation <- factor(end$gen)
mid = filter(tor_ot_mvc, diagnostic == 'contradictory_objectives' & gen == 40000)
mid$Generation <- factor(mid$gen)
mvc_p = ggplot(mid, aes(x = T, y=uni_str_pos, group = T, shape = Generation)) +
geom_point(col = mvc_col[1] , position = position_jitternudge(jitter.width = .03, nudge.x = -0.05), size = 2, alpha = 1.0) +
geom_boxplot(position = position_nudge(x = -.15, y = 0), lwd = 0.7, col = mvc_col[1], fill = mvc_col[1], width = .1, outlier.shape = NA, alpha = 0.0) +
geom_point(data = end, aes(x = T, y=uni_str_pos), col = mvc_col[2], position = position_jitternudge(jitter.width = .03, nudge.x = 0.05), size = 2, alpha = 1.0) +
geom_boxplot(data = end, aes(x = T, y=uni_str_pos), position = position_nudge(x = .15, y = 0), lwd = 0.7, col = mvc_col[2], fill = mvc_col[2], width = .1, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Coverage",
) +
scale_x_discrete(
name="T"
)+
scale_shape_manual(values=c(0,1))+
scale_colour_manual(values = c(mvc_col[1],mvc_col[2])) +
p_theme
plot_grid(
mvc_p +
ggtitle("Satisfactory trait coverage") +
theme(legend.position="none"),
legend,
nrow=2,
rel_heights = c(1,.05),
label_size = TSIZE
)
7.3.3.4.1 Stats
Summary statistics for the activation gene coverage at 40,000 and 50,000 generations.
slices = filter(tor_ot_mvc, diagnostic == 'contradictory_objectives' & (gen == 50000 | gen == 40000))
slices$Generation <- factor(slices$gen, levels = c(50000,40000))
slices %>%
group_by(T, Generation) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(uni_str_pos)),
min = min(uni_str_pos, na.rm = TRUE),
median = median(uni_str_pos, na.rm = TRUE),
mean = mean(uni_str_pos, na.rm = TRUE),
max = max(uni_str_pos, na.rm = TRUE),
IQR = IQR(uni_str_pos, na.rm = TRUE)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.## # A tibble: 16 x 9
## # Groups: T [8]
## T Generation count na_cnt min median mean max IQR
## <fct> <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 2 50000 50 0 2 5 4.6 9 2
## 2 2 40000 50 0 1 5 4.7 11 2.75
## 3 4 50000 50 0 1 5 4.86 9 2
## 4 4 40000 50 0 1 5 4.68 10 1.75
## 5 8 50000 50 0 1 4 4.64 9 2
## 6 8 40000 50 0 2 4 4.54 10 2.75
## 7 16 50000 50 0 1 5 4.92 9 2
## 8 16 40000 50 0 2 5 4.74 8 2.75
## 9 32 50000 50 0 1 5 4.86 8 2
## 10 32 40000 50 0 2 4 4.6 10 3
## 11 64 50000 50 0 2 4.5 4.74 11 3
## 12 64 40000 50 0 2 4 4.34 9 2
## 13 128 50000 50 0 1 4 4.52 11 2.75
## 14 128 40000 50 0 2 5 4.78 8 2
## 15 256 50000 50 0 2 5 4.92 9 2
## 16 256 40000 50 0 2 5 4.78 11 2T 2
wilcox.test(x = filter(slices, T == 2 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 2 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 2 & Generation == 50000)$uni_str_pos and filter(slices, T == 2 & Generation == 40000)$uni_str_pos
## W = 1205, p-value = 0.7547
## alternative hypothesis: true location shift is not equal to 0T 4
wilcox.test(x = filter(slices, T == 4 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 4 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 4 & Generation == 50000)$uni_str_pos and filter(slices, T == 4 & Generation == 40000)$uni_str_pos
## W = 1355, p-value = 0.4625
## alternative hypothesis: true location shift is not equal to 0T 8
wilcox.test(x = filter(slices, T == 8 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 8 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 8 & Generation == 50000)$uni_str_pos and filter(slices, T == 8 & Generation == 40000)$uni_str_pos
## W = 1351.5, p-value = 0.4781
## alternative hypothesis: true location shift is not equal to 0T 16
wilcox.test(x = filter(slices, T == 16 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 16 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 16 & Generation == 50000)$uni_str_pos and filter(slices, T == 16 & Generation == 40000)$uni_str_pos
## W = 1310, p-value = 0.677
## alternative hypothesis: true location shift is not equal to 0T 32
wilcox.test(x = filter(slices, T == 32 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 32 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 32 & Generation == 50000)$uni_str_pos and filter(slices, T == 32 & Generation == 40000)$uni_str_pos
## W = 1383, p-value = 0.3552
## alternative hypothesis: true location shift is not equal to 0T 64
wilcox.test(x = filter(slices, T == 64 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 64 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 64 & Generation == 50000)$uni_str_pos and filter(slices, T == 64 & Generation == 40000)$uni_str_pos
## W = 1373.5, p-value = 0.3871
## alternative hypothesis: true location shift is not equal to 0T 128
wilcox.test(x = filter(slices, T == 128 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 128 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 128 & Generation == 50000)$uni_str_pos and filter(slices, T == 128 & Generation == 40000)$uni_str_pos
## W = 1078, p-value = 0.2303
## alternative hypothesis: true location shift is not equal to 0T 256
wilcox.test(x = filter(slices, T == 256 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 256 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 256 & Generation == 50000)$uni_str_pos and filter(slices, T == 256 & Generation == 40000)$uni_str_pos
## W = 1280, p-value = 0.8358
## alternative hypothesis: true location shift is not equal to 07.4 Multi-path exploration results
Here we present the results for best performances and activation gene coverage found by each tournament selection size value replicate on the multi-path exploration diagnostic. Best performance found refers to the largest average trait score found in a given population, while activation gene coverage refers to the count of unique activation genes in the population. Note that both values fall between 0 and 100.
7.4.1 Performance
Here we analyze the performances for each parameter replicate on the multi-path exploration diagnostic.
7.4.1.1 Performance over time
Performance over time.
lines = filter(tor_ot, diagnostic == 'multipath_exploration') %>%
group_by(T, gen) %>%
dplyr::summarise(
min = min(pop_fit_max),
mean = mean(pop_fit_max),
max = max(pop_fit_max)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.ggplot(lines, aes(x=gen, y=mean / DIMENSIONALITY, group = T, fill = T, color = T, shape = T)) +
geom_ribbon(aes(ymin = min / DIMENSIONALITY, ymax = max / DIMENSIONALITY), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score",
limits=c(-1, 101),
breaks=seq(0,100, 20),
labels=c("0", "20", "40", "60", "80", "100")
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Best performance over time") +
p_theme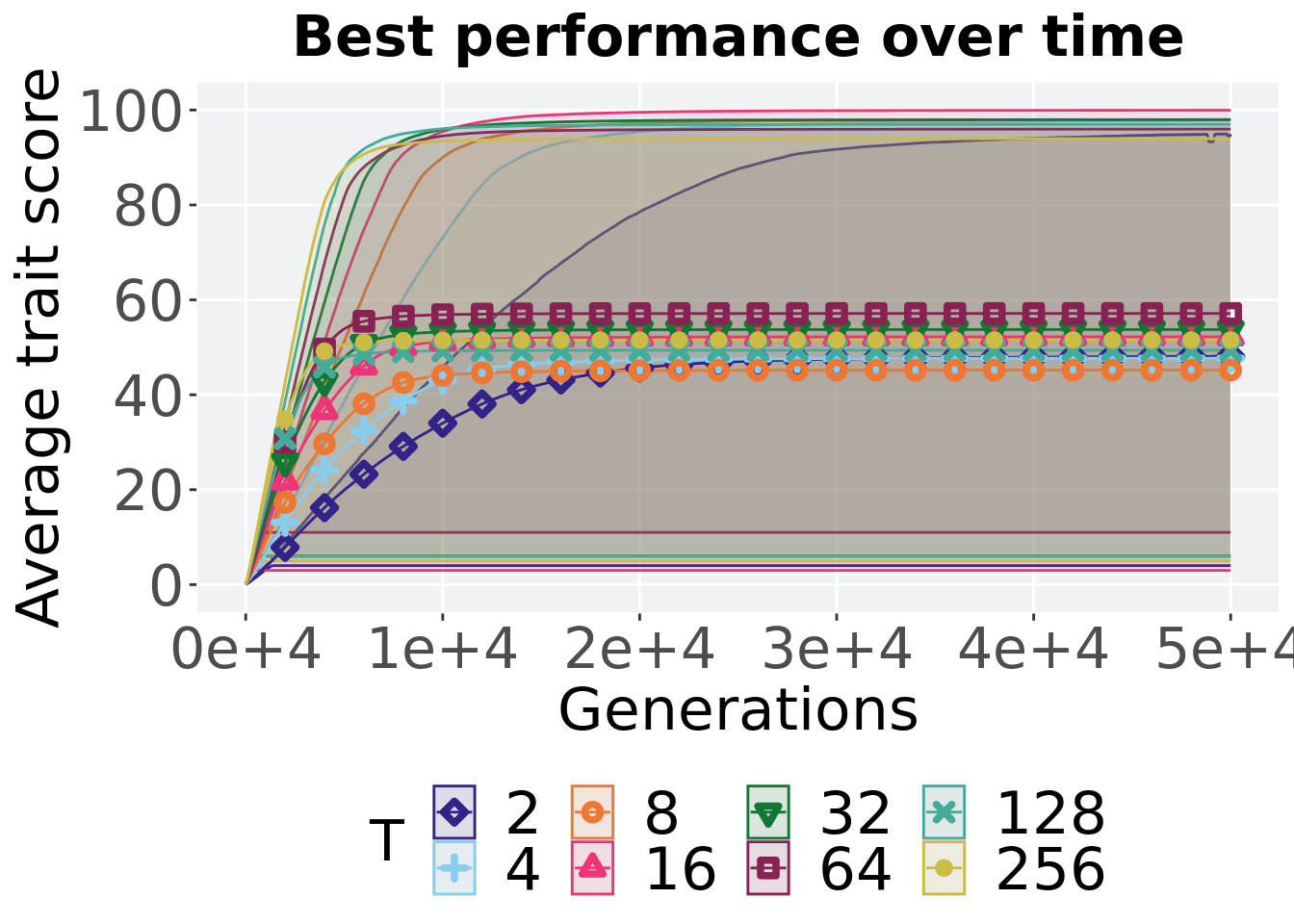
7.4.1.2 Best performance throughout
Here we plot the performance of the best performing solution found throughout 50,000 generations.
performance = filter(tor_best, col == 'pop_fit_max' & diagnostic == 'multipath_exploration') %>%
ggplot(., aes(x = T, y = val / DIMENSIONALITY, color = T, fill = T, shape = T)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Average trait score",
limits=c(-1, 101),
breaks=seq(0,100, 20),
labels=c("0", "20", "40", "60", "80", "100")
) +
scale_x_discrete(
name="T"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
p_theme7.4.1.2.1 Stats
Summary statistics for the performance of the best performing solution found throughout 50,000 generations.
performance = filter(tor_best, col == 'pop_fit_max' & diagnostic == 'multipath_exploration')
group_by(performance, T) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(val)),
min = min(val / DIMENSIONALITY, na.rm = TRUE),
median = median(val / DIMENSIONALITY, na.rm = TRUE),
mean = mean(val / DIMENSIONALITY, na.rm = TRUE),
max = max(val / DIMENSIONALITY, na.rm = TRUE),
IQR = IQR(val / DIMENSIONALITY, na.rm = TRUE)
)## # A tibble: 8 x 8
## T count na_cnt min median mean max IQR
## <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2 50 0 4 52.5 48.1 95.0 43.4
## 2 4 50 0 5 45.0 47.7 97.8 58.2
## 3 8 50 0 6.00 42.5 45.2 97.9 53.2
## 4 16 50 0 3 48.0 52.2 100. 61.5
## 5 32 50 0 6 53.0 53.8 98.0 55.2
## 6 64 50 0 11 58.0 57.1 96.0 44.0
## 7 128 50 0 6.00 52.0 49.4 97.0 47.0
## 8 256 50 0 5 51.0 51.5 94.0 41.5Kruskal–Wallis test provides evidence of no statistical differences among the best performing solution found throughout 50,000 generations.
##
## Kruskal-Wallis rank sum test
##
## data: val by T
## Kruskal-Wallis chi-squared = 6.9356, df = 7, p-value = 0.43567.4.1.3 End of 50,000 generations
Best performance in the population at the end of 50,000 generations.
filter(tor_ot, diagnostic == 'multipath_exploration' & gen == 50000) %>%
ggplot(., aes(x = T, y = pop_fit_max / DIMENSIONALITY, color = T, fill = T, shape = T)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Average trait score",
limits=c(-1, 101),
breaks=seq(0,100, 20),
labels=c("0", "20", "40", "60", "80", "100")
) +
scale_x_discrete(
name="T"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Final performance") +
p_theme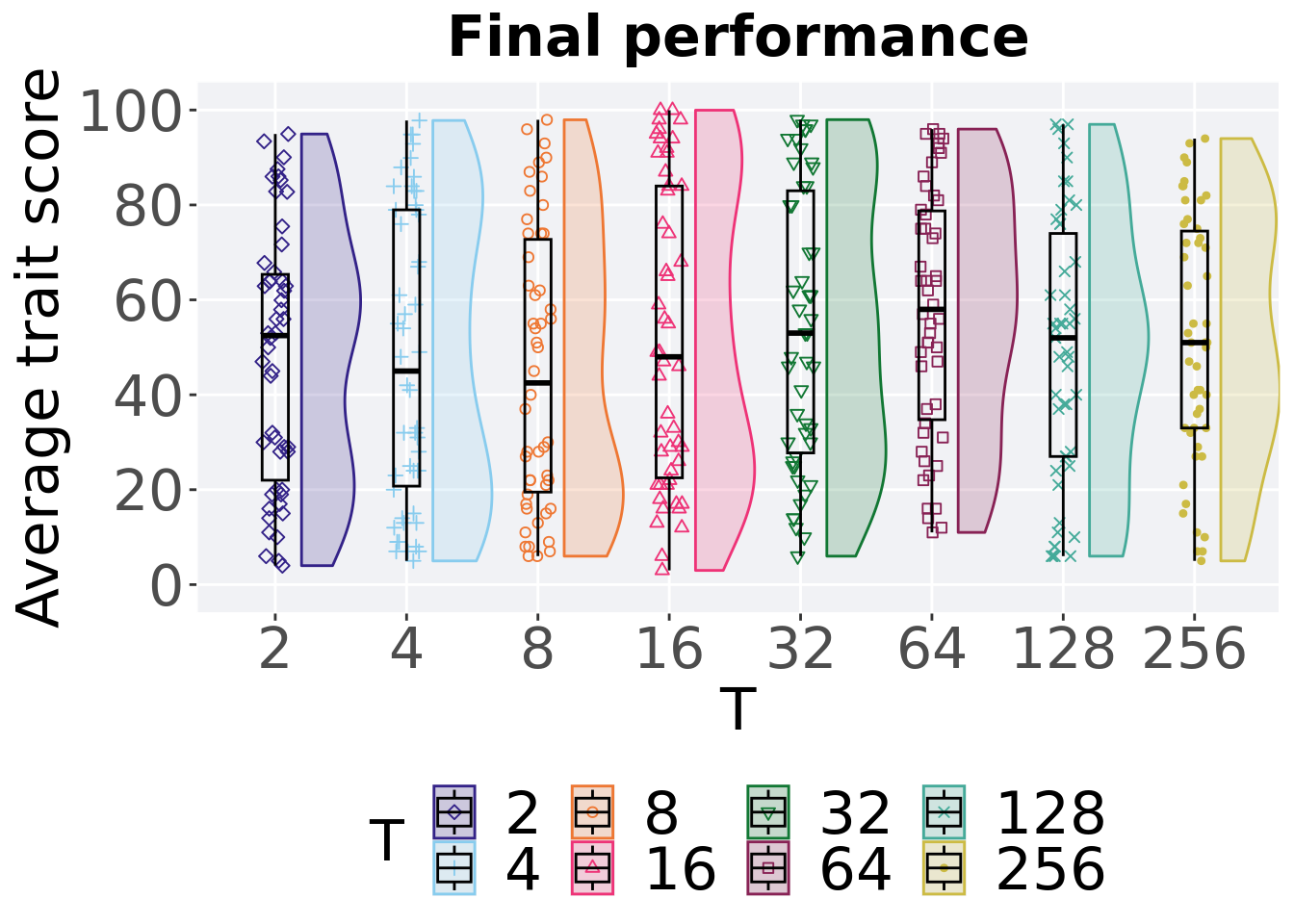
7.4.1.3.1 Stats
Summary statistics for the best performance in the population at the end of 50,000 generations.
performance = filter(tor_ot, diagnostic == 'multipath_exploration' & gen == 50000)
group_by(performance, T) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_fit_max / DIMENSIONALITY)),
min = min(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
median = median(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
mean = mean(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
max = max(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
IQR = IQR(pop_fit_max / DIMENSIONALITY, na.rm = TRUE)
)## # A tibble: 8 x 8
## T count na_cnt min median mean max IQR
## <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2 50 0 4 52.5 48.1 94.9 43.4
## 2 4 50 0 5 45.0 47.7 97.8 58.2
## 3 8 50 0 6.00 42.5 45.2 97.9 53.2
## 4 16 50 0 3 48.0 52.2 100. 61.5
## 5 32 50 0 6 53.0 53.8 98.0 55.2
## 6 64 50 0 11 58.0 57.1 96.0 44.0
## 7 128 50 0 6.00 52.0 49.4 97.0 47.0
## 8 256 50 0 5 51.0 51.5 94.0 41.5Kruskal–Wallis test provides evidence of statistical differences among best performance in the population at the end of 50,000 generations.
##
## Kruskal-Wallis rank sum test
##
## data: pop_fit_max by T
## Kruskal-Wallis chi-squared = 6.9356, df = 7, p-value = 0.4356Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction on the best performance in the population at the end of 50,000 generations.
pairwise.wilcox.test(x = performance$pop_fit_max, g = performance$T , p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'l')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: performance$pop_fit_max and performance$T
##
## 2 4 8 16 32 64 128
## 4 1 - - - - - -
## 8 1 1 - - - - -
## 16 1 1 1 - - - -
## 32 1 1 1 1 - - -
## 64 1 1 1 1 1 - -
## 128 1 1 1 1 1 1 -
## 256 1 1 1 1 1 1 1
##
## P value adjustment method: bonferroni7.4.2 Activation gene coverage
Here we analyze the activation gene coverage for each parameter replicate on the multi-path exploration diagnostic.
7.4.2.1 Coverage over time
Activation gene coverage over time.
lines = filter(tor_ot, diagnostic == 'multipath_exploration') %>%
group_by(T, gen) %>%
dplyr::summarise(
min = min(uni_str_pos),
mean = mean(uni_str_pos),
max = max(uni_str_pos)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.ggplot(lines, aes(x=gen, y=mean, group = T, fill =T, color = T, shape = T)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage",
limits=c(-1, 101),
breaks=seq(0,100, 20),
labels=c("0", "20", "40", "60", "80", "100")
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Activation gene coverage over time") +
p_theme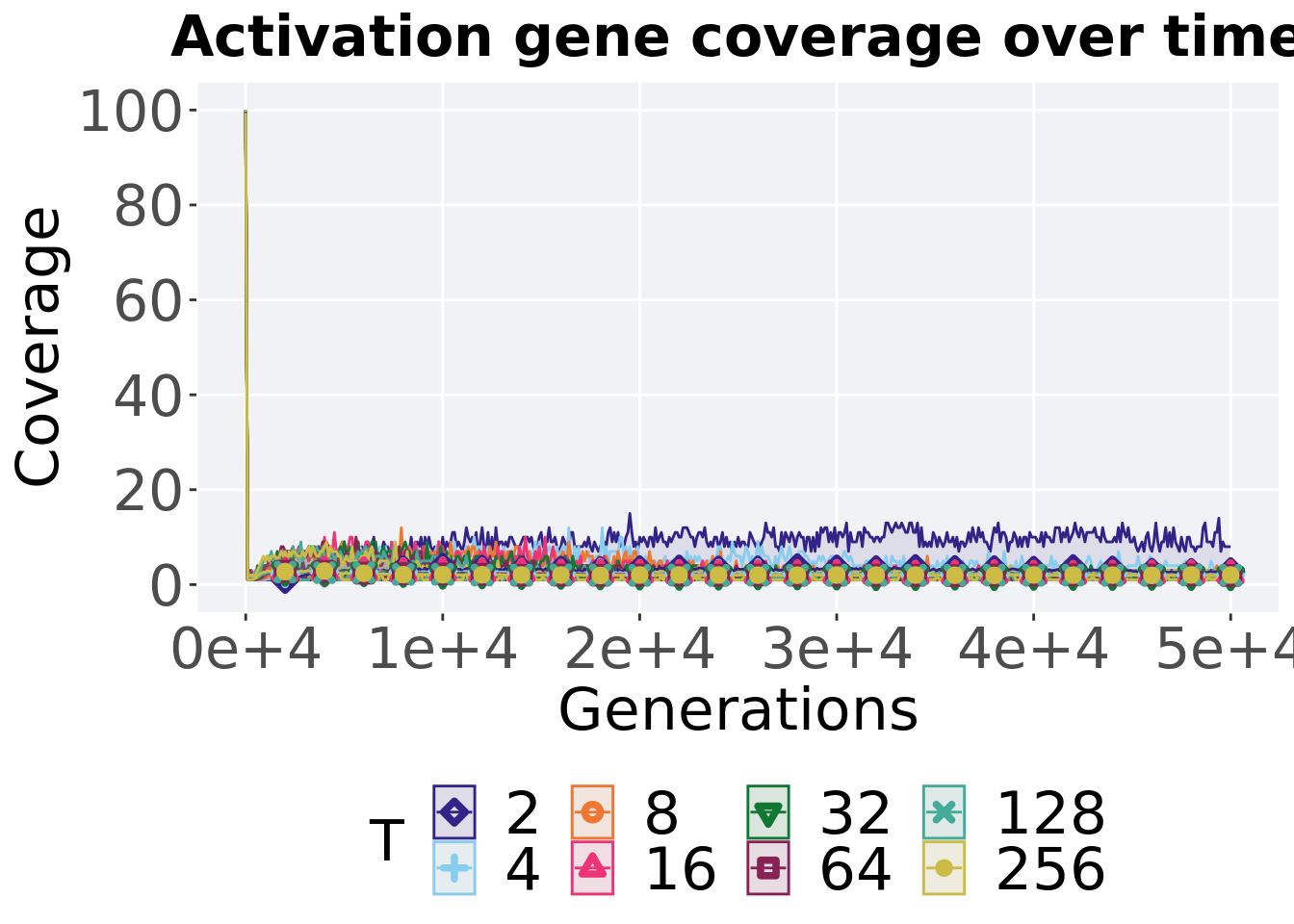
7.4.2.2 End of 50,000 generations
Activation gene coverage in the population at the end of 50,000 generations.
filter(tor_ot, diagnostic == 'multipath_exploration' & gen == 50000) %>%
ggplot(., aes(x = T, y = uni_str_pos, color = T, fill = T, shape = T)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), scale = 'width', alpha = 0.2) +
geom_point(position = position_jitter(width = .1), size = 1.5, alpha = 1.0) +
geom_boxplot(color = 'black', width = .2, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Coverage"
) +
scale_x_discrete(
name="T"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Final activation gene coverage") +
p_theme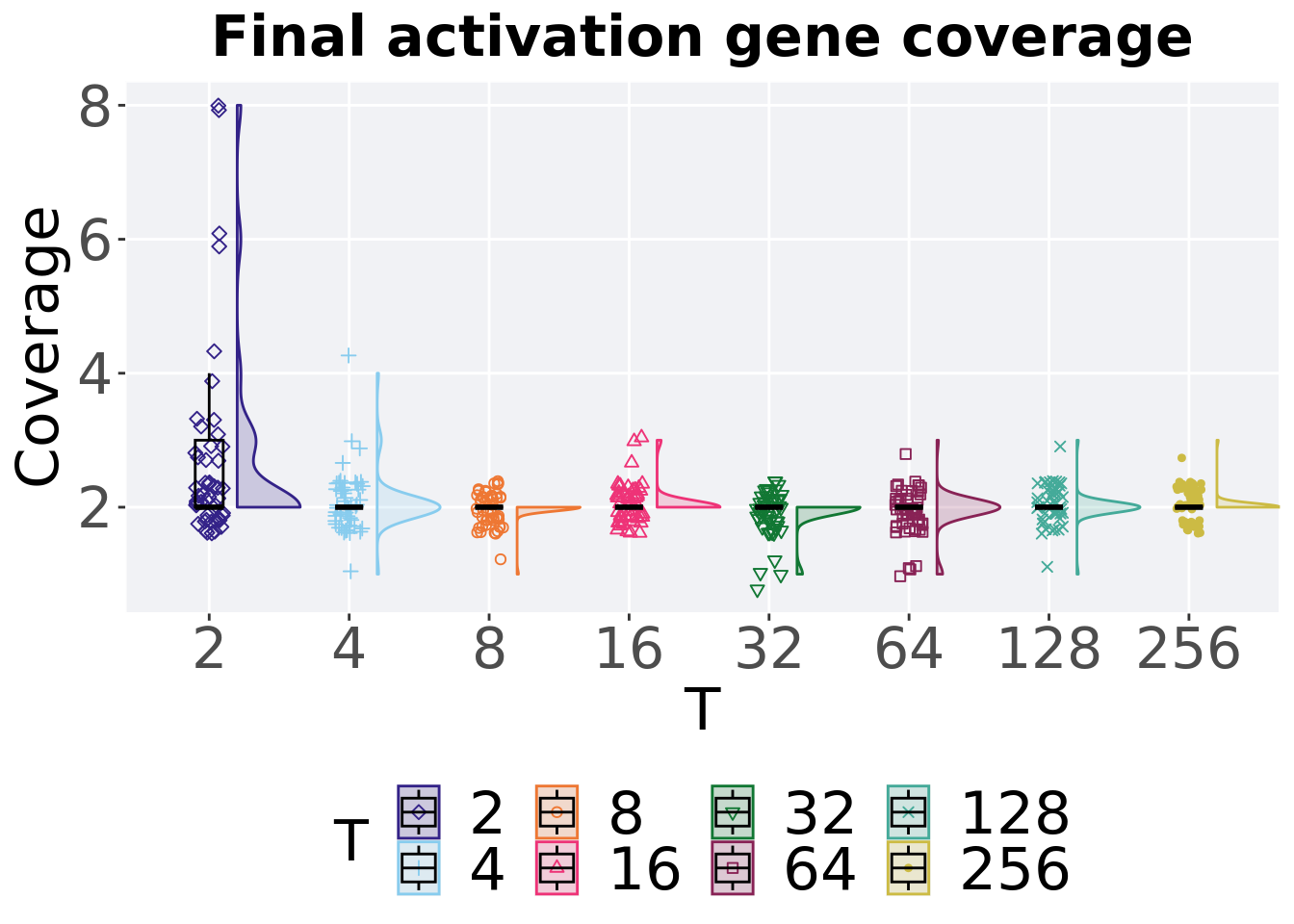
7.4.2.2.1 Stats
Summary statistics for the activation gene coverage in the population at the end of 50,000 generations.
coverage = filter(tor_ot, diagnostic == 'multipath_exploration' & gen == 50000)
group_by(coverage, T) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(uni_str_pos)),
min = min(uni_str_pos, na.rm = TRUE),
median = median(uni_str_pos, na.rm = TRUE),
mean = mean(uni_str_pos, na.rm = TRUE),
max = max(uni_str_pos, na.rm = TRUE),
IQR = IQR(uni_str_pos, na.rm = TRUE)
)## # A tibble: 8 x 8
## T count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 2 50 0 2 2 2.68 8 1
## 2 4 50 0 1 2 2.08 4 0
## 3 8 50 0 1 2 1.98 2 0
## 4 16 50 0 2 2 2.06 3 0
## 5 32 50 0 1 2 1.92 2 0
## 6 64 50 0 1 2 1.94 3 0
## 7 128 50 0 1 2 2 3 0
## 8 256 50 0 2 2 2.02 3 0Kruskal–Wallis test provides evidence of statistical differences among activation gene coverage in the population at the end of 50,000 generations.
##
## Kruskal-Wallis rank sum test
##
## data: uni_str_pos by T
## Kruskal-Wallis chi-squared = 61.183, df = 7, p-value = 8.757e-11Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction on the activation gene coverage in the population at the end of 50,000 generations.
pairwise.wilcox.test(x = coverage$uni_str_pos, g = coverage$T , p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 't')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: coverage$uni_str_pos and coverage$T
##
## 2 4 8 16 32 64 128
## 4 0.04907 - - - - - -
## 8 0.00031 1.00000 - - - - -
## 16 0.02131 1.00000 1.00000 - - - -
## 32 0.00011 0.58051 1.00000 0.24142 - - -
## 64 0.00044 1.00000 1.00000 0.99216 1.00000 - -
## 128 0.00134 1.00000 1.00000 1.00000 1.00000 1.00000 -
## 256 0.00191 1.00000 1.00000 1.00000 0.70887 1.00000 1.00000
##
## P value adjustment method: bonferroni7.4.3 Multi-valley crossing
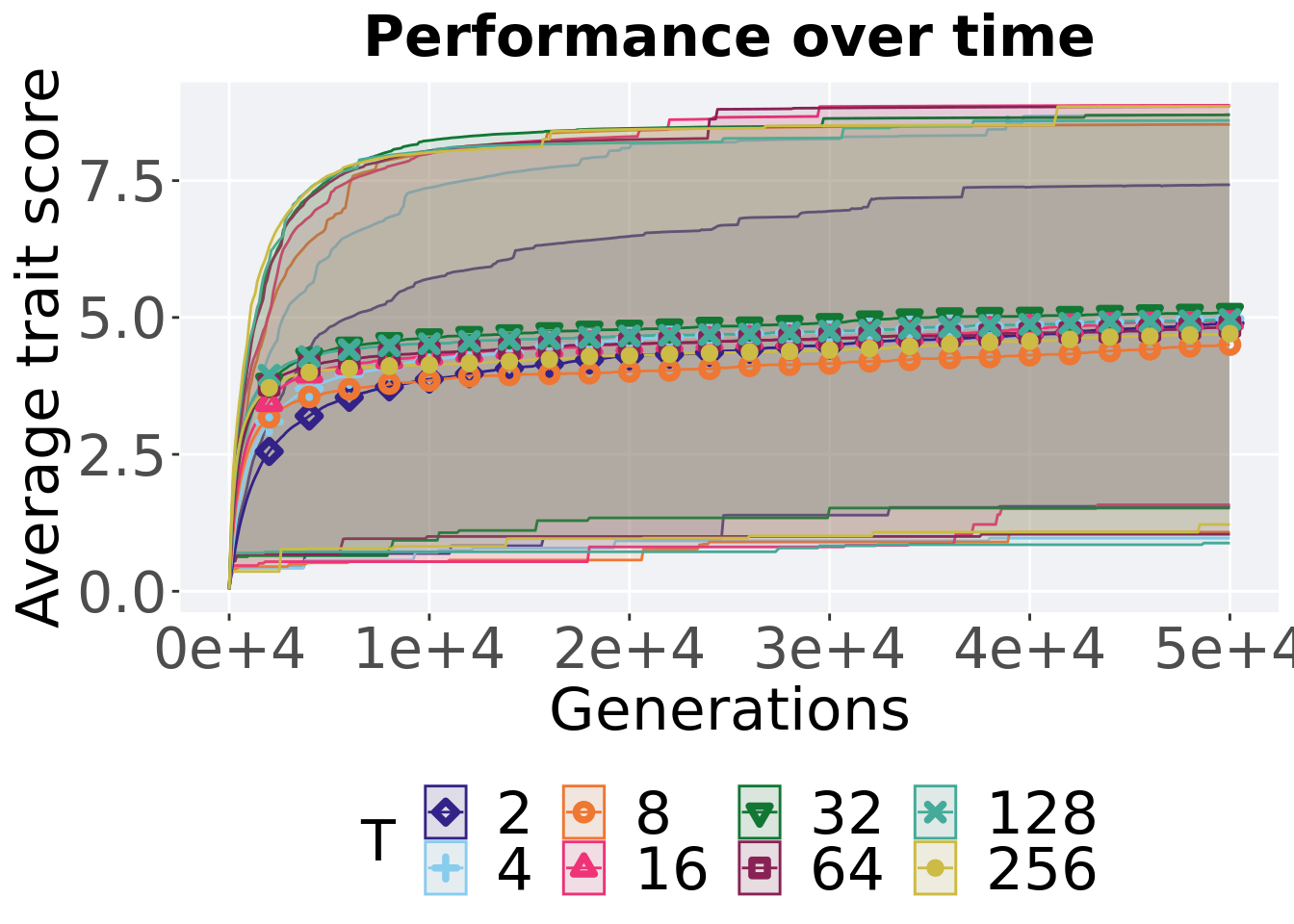
7.4.3.1 Performance over time
# data for lines and shading on plots
lines = filter(tor_ot_mvc, diagnostic == 'multipath_exploration') %>%
group_by(T, gen) %>%
dplyr::summarise(
min = min(pop_fit_max) / DIMENSIONALITY,
mean = mean(pop_fit_max) / DIMENSIONALITY,
max = max(pop_fit_max) / DIMENSIONALITY
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.ggplot(lines, aes(x=gen, y=mean, group = T, fill =T, color = T, shape = T)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Performance over time')+
p_theme +
guides(
shape=guide_legend(nrow=2, title.position = "left"),
color=guide_legend(nrow=2, title.position = "left"),
fill=guide_legend(nrow=2, title.position = "left")
)
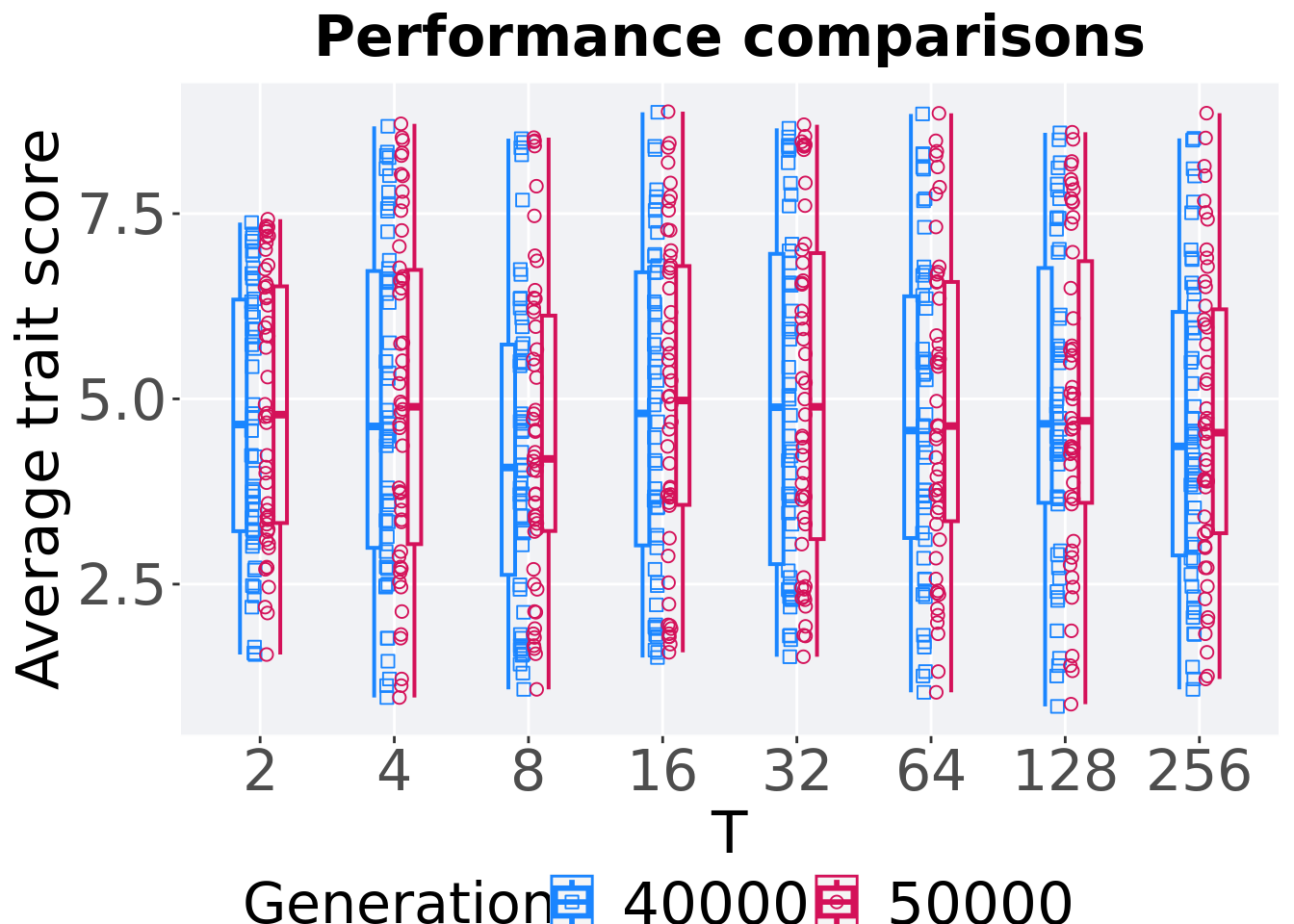
7.4.3.2 Performance comparison
Best performances in the population at 40,000 and 50,000 generations.
# 80% and final generation comparison
end = filter(tor_ot_mvc, diagnostic == 'multipath_exploration' & gen == 50000 & T != 'ran')
end$Generation <- factor(end$gen)
mid = filter(tor_ot_mvc, diagnostic == 'multipath_exploration' & gen == 40000 & T != 'ran')
mid$Generation <- factor(mid$gen)
mvc_p = ggplot(mid, aes(x = T, y=pop_fit_max / DIMENSIONALITY, group = T, shape = Generation)) +
geom_point(col = mvc_col[1] , position = position_jitternudge(jitter.width = .03, nudge.x = -0.05), size = 2, alpha = 1.0) +
geom_boxplot(position = position_nudge(x = -.15, y = 0), lwd = 0.7, col = mvc_col[1], fill = mvc_col[1], width = .1, outlier.shape = NA, alpha = 0.0) +
geom_point(data = end, aes(x = T, y=pop_fit_max / DIMENSIONALITY), col = mvc_col[2], position = position_jitternudge(jitter.width = .03, nudge.x = 0.05), size = 2, alpha = 1.0) +
geom_boxplot(data = end, aes(x = T, y=pop_fit_max / DIMENSIONALITY), position = position_nudge(x = .15, y = 0), lwd = 0.7, col = mvc_col[2], fill = mvc_col[2], width = .1, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_discrete(
name="T"
)+
scale_shape_manual(values=c(0,1))+
scale_colour_manual(values = c(mvc_col[1],mvc_col[2])) +
p_theme
plot_grid(
mvc_p +
ggtitle("Performance comparisons") +
theme(legend.position="none"),
legend,
nrow=2,
rel_heights = c(1,.05),
label_size = TSIZE
)
7.4.3.3 Stats
Summary statistics for the performance of the best performance at 40,000 and 50,000 generations.
slices = filter(tor_ot_mvc, diagnostic == 'multipath_exploration' & (gen == 50000 | gen == 40000))
slices$Generation <- factor(slices$gen, levels = c(50000,40000))
slices %>%
group_by(T, Generation) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_fit_max / DIMENSIONALITY)),
min = min(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
median = median(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
mean = mean(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
max = max(pop_fit_max / DIMENSIONALITY, na.rm = TRUE),
IQR = IQR(pop_fit_max / DIMENSIONALITY, na.rm = TRUE)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.## # A tibble: 16 x 9
## # Groups: T [8]
## T Generation count na_cnt min median mean max IQR
## <fct> <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2 50000 50 0 1.55 4.79 4.89 7.43 3.19
## 2 2 40000 50 0 1.55 4.65 4.68 7.38 3.13
## 3 4 50000 50 0 0.970 4.89 4.98 8.71 3.70
## 4 4 40000 50 0 0.970 4.63 4.87 8.68 3.74
## 5 8 50000 50 0 1.08 4.19 4.50 8.53 2.91
## 6 8 40000 50 0 1.08 4.07 4.31 8.51 3.11
## 7 16 50000 50 0 1.58 4.98 4.94 8.88 3.22
## 8 16 40000 50 0 1.51 4.80 4.80 8.87 3.69
## 9 32 50000 50 0 1.52 4.89 5.09 8.70 3.86
## 10 32 40000 50 0 1.52 4.89 5.02 8.65 4.19
## 11 64 50000 50 0 1.04 4.63 4.84 8.85 3.23
## 12 64 40000 50 0 1.04 4.57 4.71 8.85 3.26
## 13 128 50000 50 0 0.880 4.70 4.96 8.60 3.26
## 14 128 40000 50 0 0.850 4.66 4.88 8.59 3.17
## 15 256 50000 50 0 1.22 4.54 4.70 8.86 3.02
## 16 256 40000 50 0 1.08 4.36 4.57 8.51 3.29T 2
wilcox.test(x = filter(slices, T == 2 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 2 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 2 & Generation == 50000)$pop_fit_max and filter(slices, T == 2 & Generation == 40000)$pop_fit_max
## W = 1354, p-value = 0.4755
## alternative hypothesis: true location shift is not equal to 0T 4
wilcox.test(x = filter(slices, T == 4 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 4 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 4 & Generation == 50000)$pop_fit_max and filter(slices, T == 4 & Generation == 40000)$pop_fit_max
## W = 1305, p-value = 0.7071
## alternative hypothesis: true location shift is not equal to 0T 8
wilcox.test(x = filter(slices, T == 8 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 8 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 8 & Generation == 50000)$pop_fit_max and filter(slices, T == 8 & Generation == 40000)$pop_fit_max
## W = 1339, p-value = 0.5418
## alternative hypothesis: true location shift is not equal to 0T 16
wilcox.test(x = filter(slices, T == 16 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 16 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 16 & Generation == 50000)$pop_fit_max and filter(slices, T == 16 & Generation == 40000)$pop_fit_max
## W = 1322, p-value = 0.6221
## alternative hypothesis: true location shift is not equal to 0T 32
wilcox.test(x = filter(slices, T == 32 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 32 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 32 & Generation == 50000)$pop_fit_max and filter(slices, T == 32 & Generation == 40000)$pop_fit_max
## W = 1298, p-value = 0.7433
## alternative hypothesis: true location shift is not equal to 0T 64
wilcox.test(x = filter(slices, T == 64 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 64 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 64 & Generation == 50000)$pop_fit_max and filter(slices, T == 64 & Generation == 40000)$pop_fit_max
## W = 1314, p-value = 0.6616
## alternative hypothesis: true location shift is not equal to 0T 128
wilcox.test(x = filter(slices, T == 128 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 128 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 128 & Generation == 50000)$pop_fit_max and filter(slices, T == 128 & Generation == 40000)$pop_fit_max
## W = 1304, p-value = 0.7123
## alternative hypothesis: true location shift is not equal to 0T 256
wilcox.test(x = filter(slices, T == 256 & Generation == 50000)$pop_fit_max,
y = filter(slices, T == 256 & Generation == 40000)$pop_fit_max,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 256 & Generation == 50000)$pop_fit_max and filter(slices, T == 256 & Generation == 40000)$pop_fit_max
## W = 1316.5, p-value = 0.6491
## alternative hypothesis: true location shift is not equal to 07.4.3.4 Activation gene coverage over time
lines = filter(tor_ot_mvc, diagnostic == 'multipath_exploration') %>%
group_by(T, gen) %>%
dplyr::summarise(
min = min(uni_str_pos),
mean = mean(uni_str_pos),
max = max(uni_str_pos)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.ggplot(lines, aes(x=gen, y=mean, group = T, fill =T, color = T, shape = T)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle("Activation gene coverage over time") +
p_theme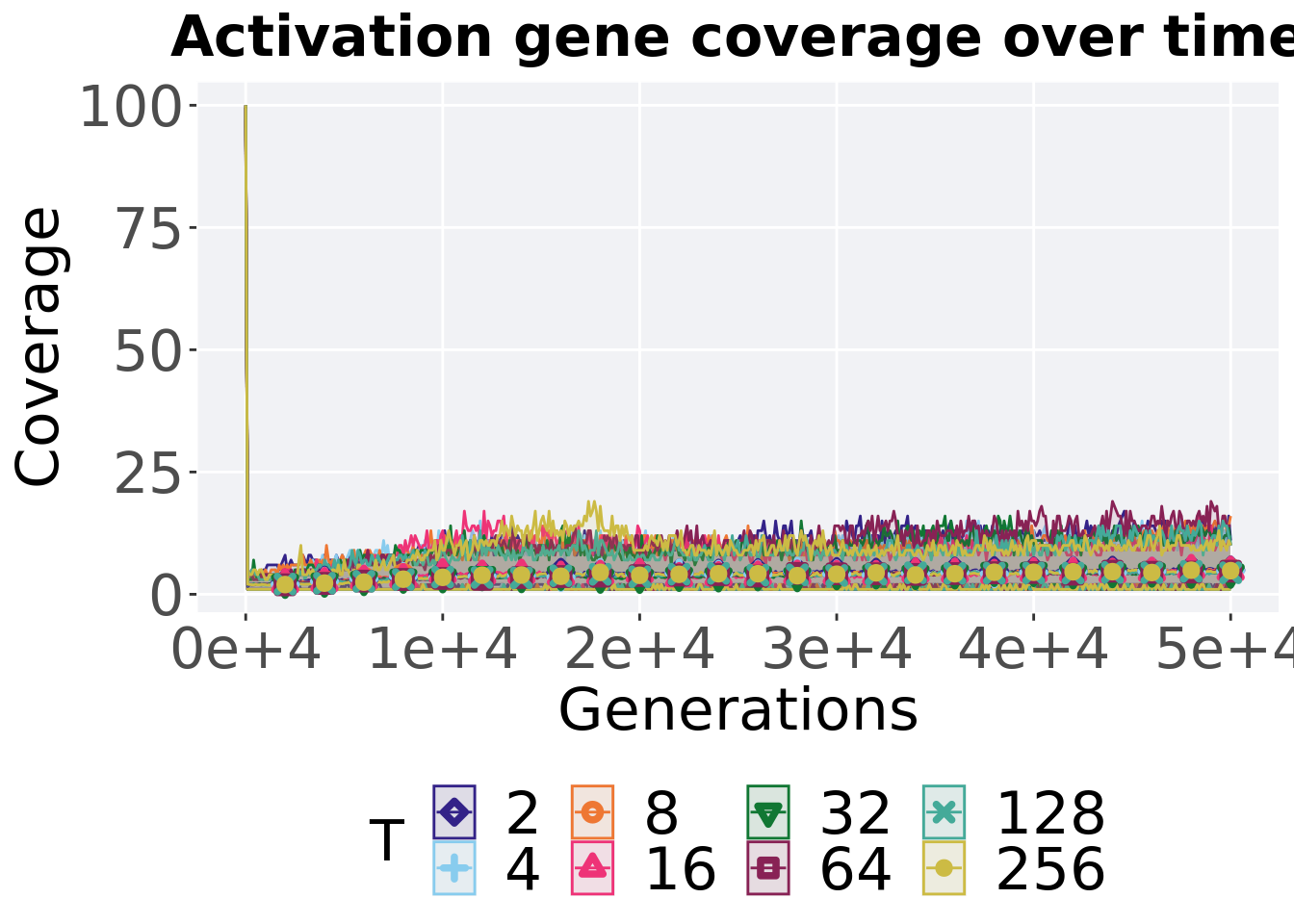
7.4.3.5 Activation gene coverage comparison
Activation gene coverage in the population at 40,000 and 50,000 generations.
# 80% and final generation comparison
end = filter(tor_ot_mvc, diagnostic == 'multipath_exploration' & gen == 50000)
end$Generation <- factor(end$gen)
mid = filter(tor_ot_mvc, diagnostic == 'multipath_exploration' & gen == 40000)
mid$Generation <- factor(mid$gen)
mvc_p = ggplot(mid, aes(x = T, y=uni_str_pos, group = T, shape = Generation)) +
geom_point(col = mvc_col[1] , position = position_jitternudge(jitter.width = .03, nudge.x = -0.05), size = 2, alpha = 1.0) +
geom_boxplot(position = position_nudge(x = -.15, y = 0), lwd = 0.7, col = mvc_col[1], fill = mvc_col[1], width = .1, outlier.shape = NA, alpha = 0.0) +
geom_point(data = end, aes(x = T, y=uni_str_pos), col = mvc_col[2], position = position_jitternudge(jitter.width = .03, nudge.x = 0.05), size = 2, alpha = 1.0) +
geom_boxplot(data = end, aes(x = T, y=uni_str_pos), position = position_nudge(x = .15, y = 0), lwd = 0.7, col = mvc_col[2], fill = mvc_col[2], width = .1, outlier.shape = NA, alpha = 0.0) +
scale_y_continuous(
name="Coverage",
) +
scale_x_discrete(
name="T"
)+
scale_shape_manual(values=c(0,1))+
scale_colour_manual(values = c(mvc_col[1],mvc_col[2])) +
p_theme
plot_grid(
mvc_p +
ggtitle("Activation gene coverage") +
theme(legend.position="none"),
legend,
nrow=2,
rel_heights = c(1,.05),
label_size = TSIZE
)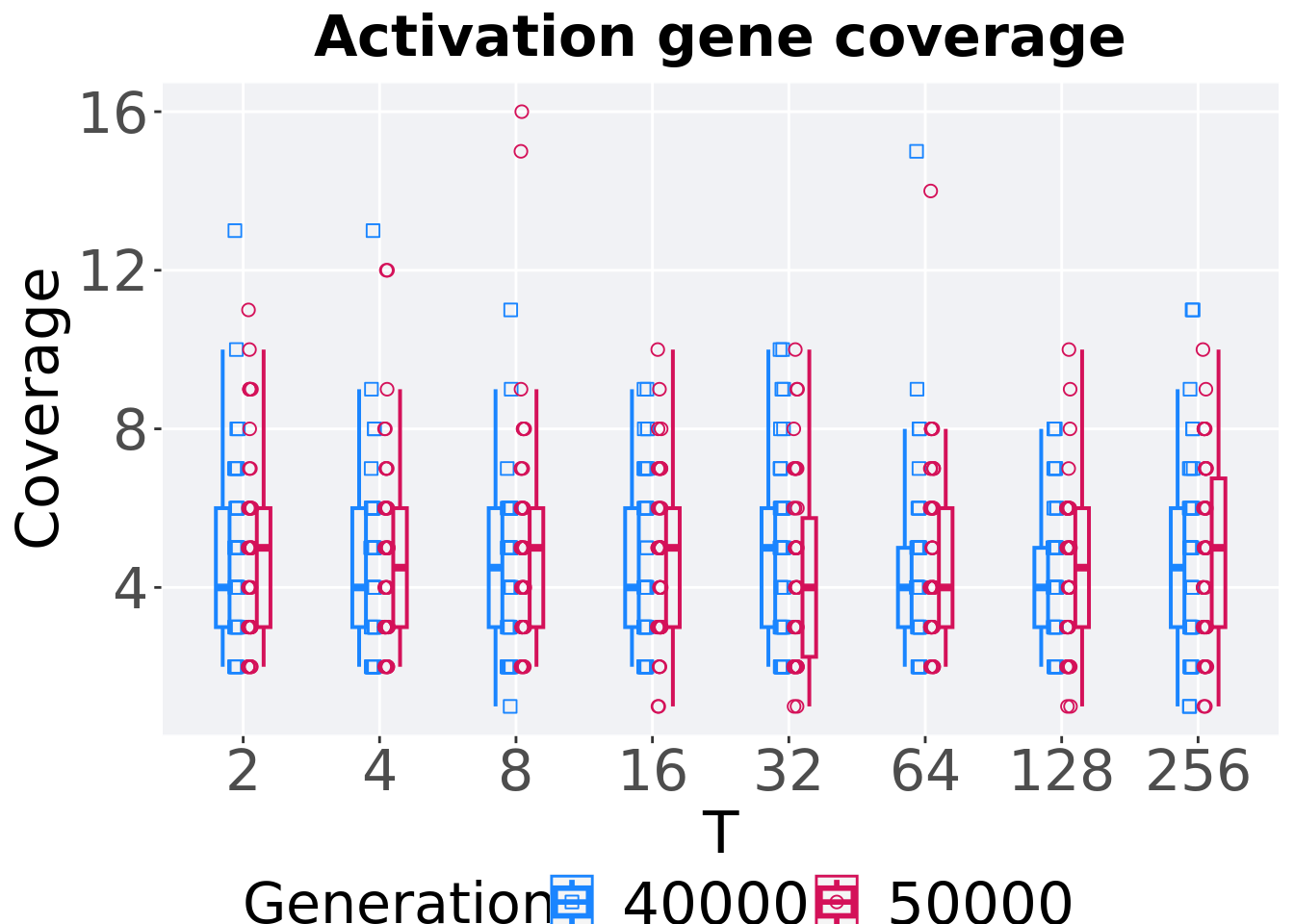
7.4.3.5.1 Stats
Summary statistics for the activation gene coverage at 40,000 and 50,000 generations.
slices = filter(tor_ot_mvc, diagnostic == 'multipath_exploration' & (gen == 50000 | gen == 40000))
slices$Generation <- factor(slices$gen, levels = c(50000,40000))
slices %>%
group_by(T, Generation) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(uni_str_pos)),
min = min(uni_str_pos, na.rm = TRUE),
median = median(uni_str_pos, na.rm = TRUE),
mean = mean(uni_str_pos, na.rm = TRUE),
max = max(uni_str_pos, na.rm = TRUE),
IQR = IQR(uni_str_pos, na.rm = TRUE)
)## `summarise()` has grouped output by 'T'. You can override using the `.groups`
## argument.## # A tibble: 16 x 9
## # Groups: T [8]
## T Generation count na_cnt min median mean max IQR
## <fct> <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 2 50000 50 0 2 5 4.94 11 3
## 2 2 40000 50 0 2 4 4.76 13 3
## 3 4 50000 50 0 2 4.5 4.8 12 3
## 4 4 40000 50 0 2 4 4.48 13 3
## 5 8 50000 50 0 2 5 5.1 16 3
## 6 8 40000 50 0 1 4.5 4.38 11 3
## 7 16 50000 50 0 1 5 4.68 10 3
## 8 16 40000 50 0 2 4 4.4 9 3
## 9 32 50000 50 0 1 4 4.34 10 3.5
## 10 32 40000 50 0 2 5 4.66 10 3
## 11 64 50000 50 0 2 4 4.72 14 3
## 12 64 40000 50 0 2 4 4.44 15 2
## 13 128 50000 50 0 1 4.5 4.4 10 3
## 14 128 40000 50 0 2 4 4.38 8 2
## 15 256 50000 50 0 1 5 4.82 10 3.75
## 16 256 40000 50 0 1 4.5 4.5 11 3T 2
wilcox.test(x = filter(slices, T == 2 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 2 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 2 & Generation == 50000)$uni_str_pos and filter(slices, T == 2 & Generation == 40000)$uni_str_pos
## W = 1272.5, p-value = 0.878
## alternative hypothesis: true location shift is not equal to 0T 4
wilcox.test(x = filter(slices, T == 4 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 4 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 4 & Generation == 50000)$uni_str_pos and filter(slices, T == 4 & Generation == 40000)$uni_str_pos
## W = 1347.5, p-value = 0.498
## alternative hypothesis: true location shift is not equal to 0T 8
wilcox.test(x = filter(slices, T == 8 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 8 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 8 & Generation == 50000)$uni_str_pos and filter(slices, T == 8 & Generation == 40000)$uni_str_pos
## W = 1396, p-value = 0.3094
## alternative hypothesis: true location shift is not equal to 0T 16
wilcox.test(x = filter(slices, T == 16 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 16 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 16 & Generation == 50000)$uni_str_pos and filter(slices, T == 16 & Generation == 40000)$uni_str_pos
## W = 1386.5, p-value = 0.3422
## alternative hypothesis: true location shift is not equal to 0T 32
wilcox.test(x = filter(slices, T == 32 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 32 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 32 & Generation == 50000)$uni_str_pos and filter(slices, T == 32 & Generation == 40000)$uni_str_pos
## W = 1155.5, p-value = 0.5116
## alternative hypothesis: true location shift is not equal to 0T 64
wilcox.test(x = filter(slices, T == 64 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 64 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 64 & Generation == 50000)$uni_str_pos and filter(slices, T == 64 & Generation == 40000)$uni_str_pos
## W = 1346.5, p-value = 0.5018
## alternative hypothesis: true location shift is not equal to 0T 128
wilcox.test(x = filter(slices, T == 128 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 128 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 128 & Generation == 50000)$uni_str_pos and filter(slices, T == 128 & Generation == 40000)$uni_str_pos
## W = 1260.5, p-value = 0.9443
## alternative hypothesis: true location shift is not equal to 0T 256
wilcox.test(x = filter(slices, T == 256 & Generation == 50000)$uni_str_pos,
y = filter(slices, T == 256 & Generation == 40000)$uni_str_pos,
alternative = 't')##
## Wilcoxon rank sum test with continuity correction
##
## data: filter(slices, T == 256 & Generation == 50000)$uni_str_pos and filter(slices, T == 256 & Generation == 40000)$uni_str_pos
## W = 1377.5, p-value = 0.3761
## alternative hypothesis: true location shift is not equal to 0