Chapter 5 Phenotypic fitness sharing
Results for the phenotypic fitness sharing parameter sweep on the diagnostics with no valleys.
5.1 Data setup
over_time_df <- read.csv(paste(DATA_DIR,'OVER-TIME/pfs.csv', sep = "", collapse = NULL), header = TRUE, stringsAsFactors = FALSE)
over_time_df$Sigma <- factor(over_time_df$Sigma, levels = FS_LIST)
best_df <- read.csv(paste(DATA_DIR,'BEST/pfs.csv', sep = "", collapse = NULL), header = TRUE, stringsAsFactors = FALSE)
best_df$Sigma <- factor(best_df$Sigma, levels = FS_LIST)
sati_df <- read.csv(paste(DATA_DIR,'SOL-FND/pfs.csv', sep = "", collapse = NULL), header = TRUE, stringsAsFactors = FALSE)
sati_df$Sigma <- factor(sati_df$Sigma, levels = FS_LIST)5.2 Exploitation rate results
Here we present the results for best performances found by each selection scheme parameter on the exploitation rate diagnostic. 50 replicates are conducted for each scheme explored.
5.2.1 Performance over time
Best performance in a population over time. Data points on the graph is the average performance across 50 replicates every 2000 generations. Shading comes from the best and worse performance across 50 replicates.
lines = filter(over_time_df, acro == 'exp') %>%
group_by(Sigma, gen) %>%
dplyr::summarise(
min = min(pop_fit_max) / DIMENSIONALITY,
mean = mean(pop_fit_max) / DIMENSIONALITY,
max = max(pop_fit_max) / DIMENSIONALITY
)## `summarise()` has grouped output by 'Sigma'. You can override using the
## `.groups` argument.over_time_plot = ggplot(lines, aes(x=gen, y=mean, group = Sigma, fill = Sigma, color = Sigma, shape = Sigma)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Performance over time')+
p_theme +
guides(
shape=guide_legend(nrow=1, title.position = "left", title = 'Sigma'),
color=guide_legend(nrow=1, title.position = "left", title = 'Sigma'),
fill=guide_legend(nrow=1, title.position = "left", title = 'Sigma')
)
over_time_plot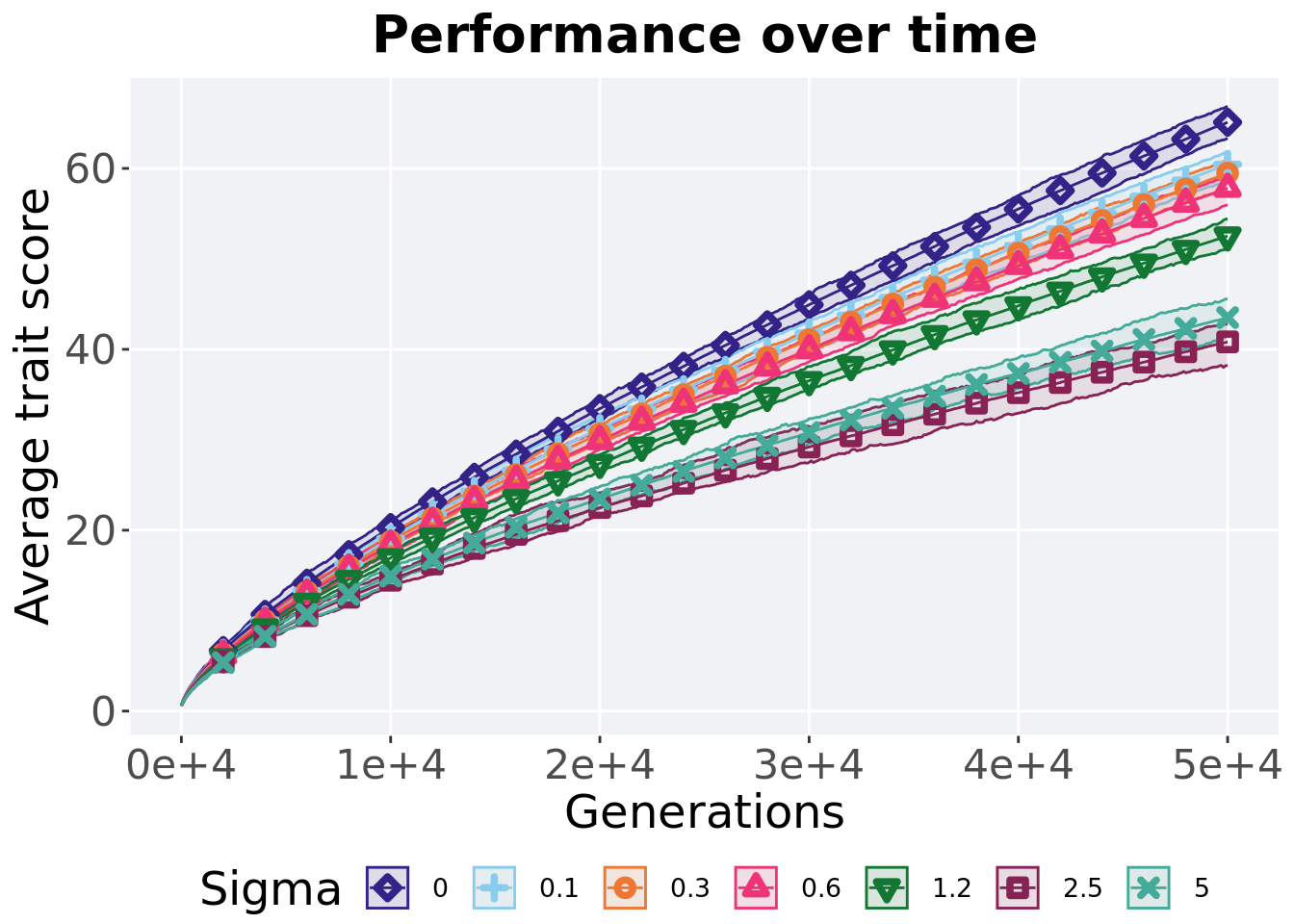
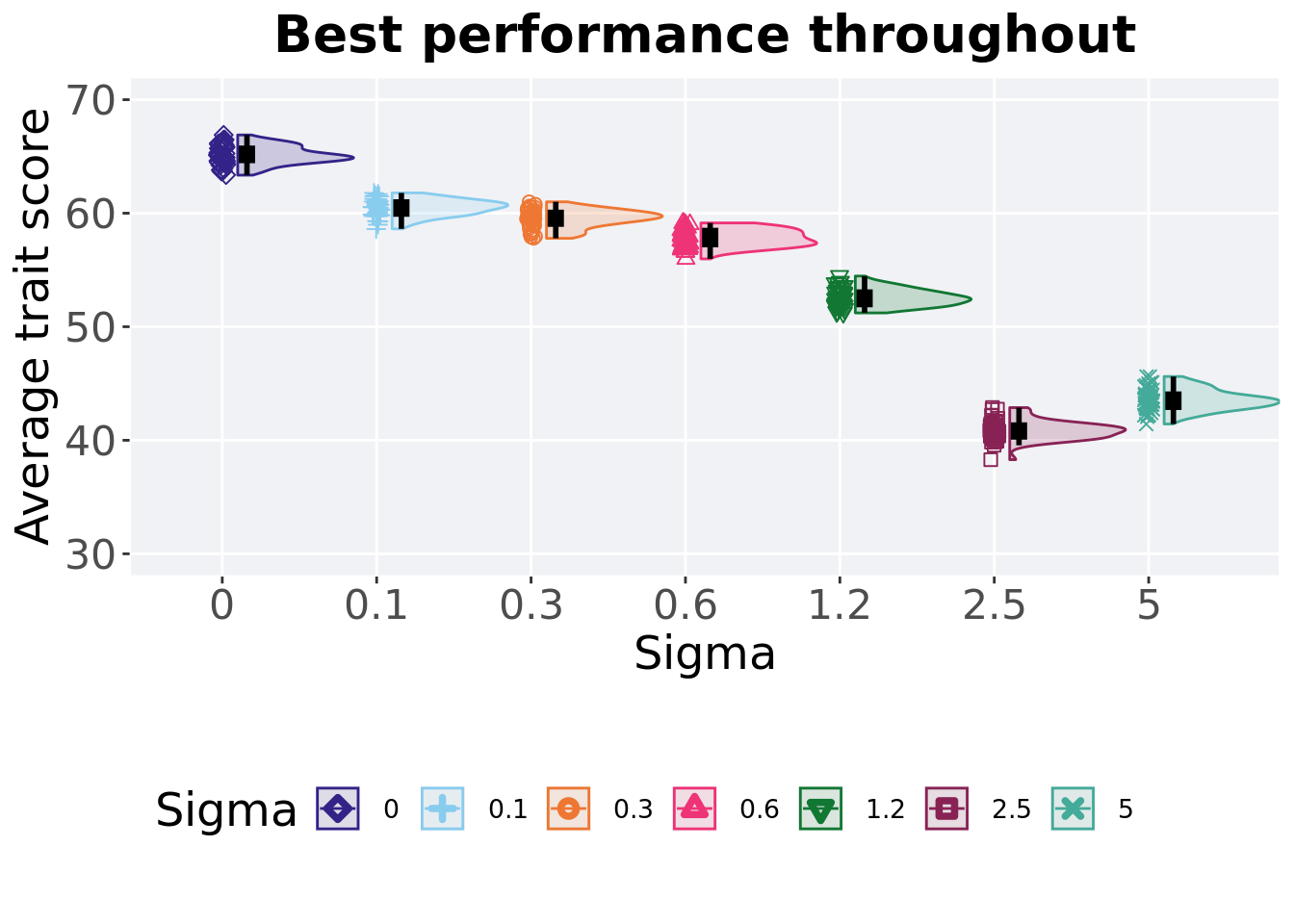
5.2.2 Best performance throughout
Best performance reached throughout 50,000 generations in a population.
plot = filter(best_df, var == 'pop_fit_max' & acro == 'exp') %>%
ggplot(., aes(x = Sigma, y = val / DIMENSIONALITY, color = Sigma, fill = Sigma, shape = Sigma)) +
geom_flat_violin(position = position_nudge(x = .1, y = 0), scale = 'width', alpha = 0.2, width = 1.5) +
geom_boxplot(color = 'black', width = .07, outlier.shape = NA, alpha = 0.0, size = 1.0, position = position_nudge(x = .16, y = 0)) +
geom_point(position = position_jitter(width = 0.03, height = 0.02), size = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score",
limits = c(30,70)
) +
scale_x_discrete(
name="Sigma"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette, ) +
scale_fill_manual(values = cb_palette) +
ggtitle('Best performance throughout')+
p_theme
plot_grid(
plot +
theme(legend.position="none"),
legend,
nrow=2,
rel_heights = c(3,1)
)
5.2.2.1 Stats
Summary statistics for the best performance.
performance = filter(best_df, var == 'pop_fit_max' & acro == 'exp')
performance$Sigma = factor(performance$Sigma, levels = c('0','0.1','0.3','0.6','1.2','5','2.5'))
performance %>%
group_by(Sigma) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(val)),
min = min(val / DIMENSIONALITY, na.rm = TRUE),
median = median(val / DIMENSIONALITY, na.rm = TRUE),
mean = mean(val / DIMENSIONALITY, na.rm = TRUE),
max = max(val / DIMENSIONALITY, na.rm = TRUE),
IQR = IQR(val / DIMENSIONALITY, na.rm = TRUE)
)## # A tibble: 7 x 8
## Sigma count na_cnt min median mean max IQR
## <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0 50 0 63.4 65.0 65.1 66.9 1.07
## 2 0.1 50 0 58.6 60.5 60.5 61.8 1.06
## 3 0.3 50 0 57.8 59.6 59.5 61.0 1.03
## 4 0.6 50 0 56.0 57.8 57.8 59.1 1.24
## 5 1.2 50 0 51.2 52.5 52.6 54.5 1.10
## 6 5 50 0 41.4 43.5 43.6 45.6 1.15
## 7 2.5 50 0 38.3 40.9 40.9 42.9 1.04Kruskal–Wallis test illustrates evidence of statistical differences.
##
## Kruskal-Wallis rank sum test
##
## data: val by Sigma
## Kruskal-Wallis chi-squared = 335.67, df = 6, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction.
pairwise.wilcox.test(x = performance$val, g = performance$Sigma, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'l')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: performance$val and performance$Sigma
##
## 0 0.1 0.3 0.6 1.2 5
## 0.1 < 2e-16 - - - - -
## 0.3 < 2e-16 6.4e-07 - - - -
## 0.6 < 2e-16 < 2e-16 1.7e-12 - - -
## 1.2 < 2e-16 < 2e-16 < 2e-16 < 2e-16 - -
## 5 < 2e-16 < 2e-16 < 2e-16 < 2e-16 < 2e-16 -
## 2.5 < 2e-16 < 2e-16 < 2e-16 < 2e-16 < 2e-16 5.9e-16
##
## P value adjustment method: bonferroni5.3 Ordered exploitation results
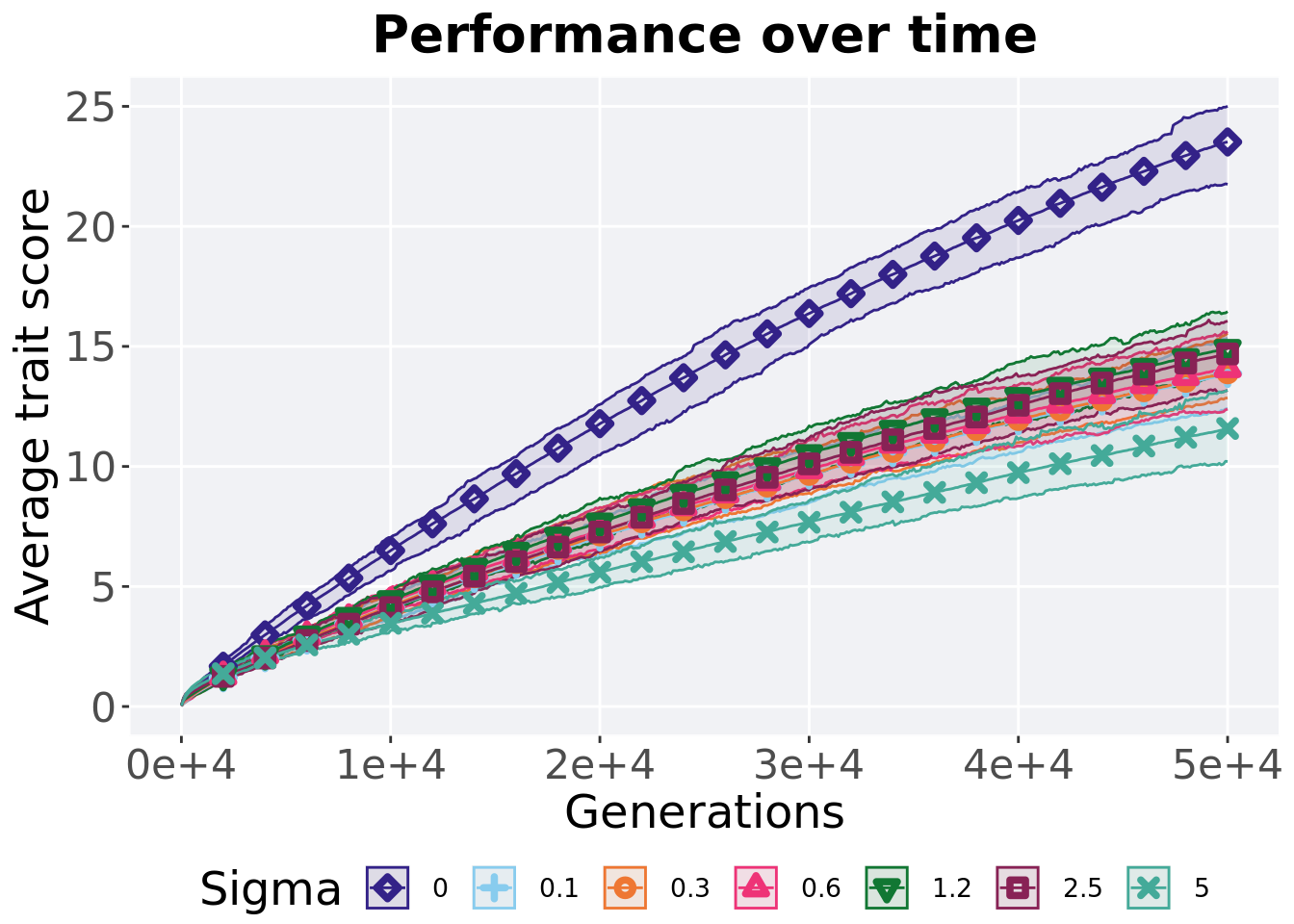
Here we present the results for best performances found by each selection scheme parameter on the exploitation rate diagnostic. 50 replicates are conducted for each scheme explored.
5.3.1 Performance over time
Best performance in a population over time. Data points on the graph is the average performance across 50 replicates every 2000 generations. Shading comes from the best and worse performance across 50 replicates.
lines = filter(over_time_df, acro == 'ord') %>%
group_by(Sigma, gen) %>%
dplyr::summarise(
min = min(pop_fit_max) / DIMENSIONALITY,
mean = mean(pop_fit_max) / DIMENSIONALITY,
max = max(pop_fit_max) / DIMENSIONALITY
)## `summarise()` has grouped output by 'Sigma'. You can override using the
## `.groups` argument.ggplot(lines, aes(x=gen, y=mean, group = Sigma, fill = Sigma, color = Sigma, shape = Sigma)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Performance over time')+
p_theme +
guides(
shape=guide_legend(nrow=1, title.position = "left", title = 'Sigma'),
color=guide_legend(nrow=1, title.position = "left", title = 'Sigma'),
fill=guide_legend(nrow=1, title.position = "left", title = 'Sigma')
)
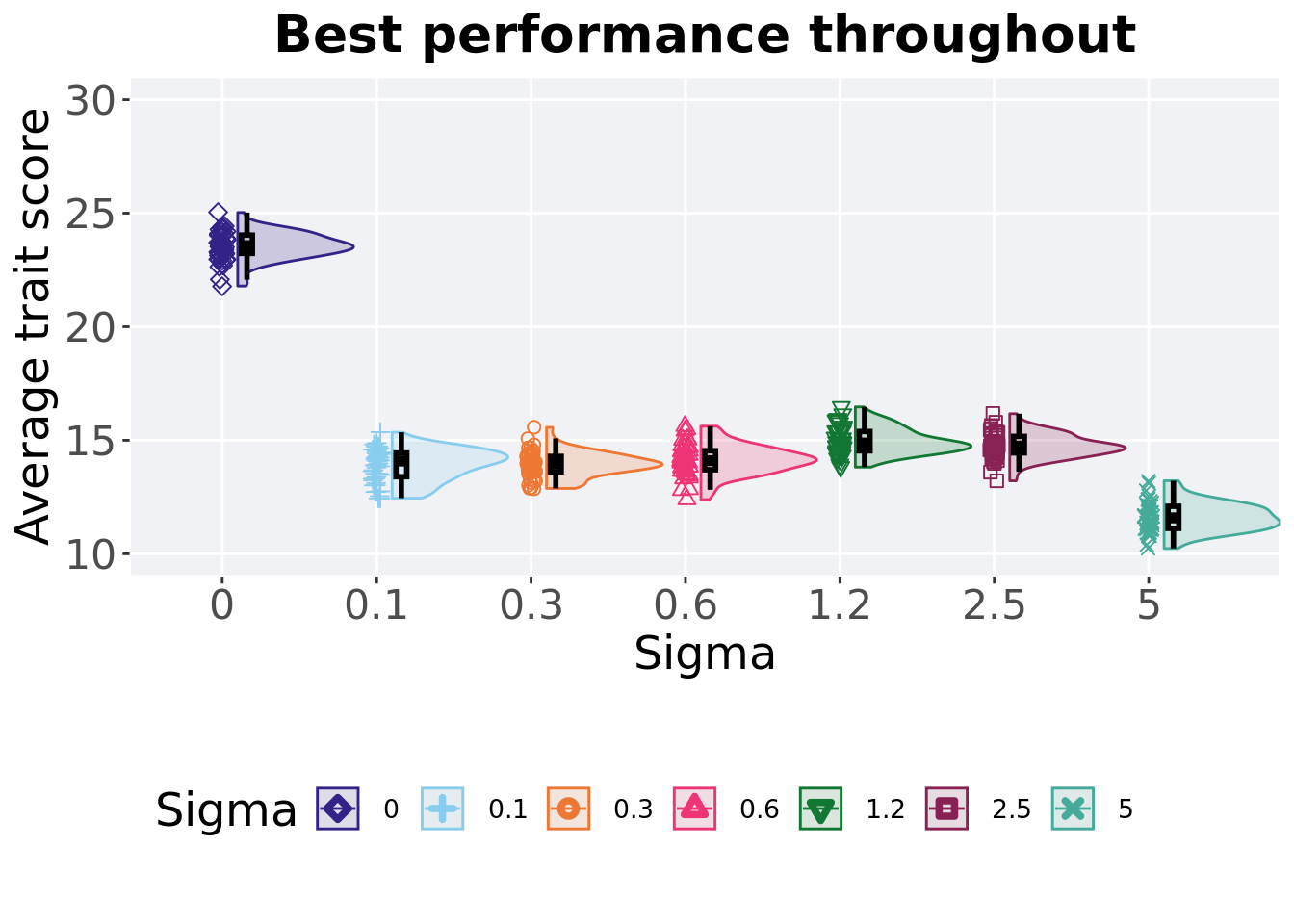
5.3.2 Best performance throughout
Best performance reached throughout 50,000 generations in a population.
plot = filter(best_df, var == 'pop_fit_max' & acro == 'ord') %>%
ggplot(., aes(x = Sigma, y = val / DIMENSIONALITY, color = Sigma, fill = Sigma, shape = Sigma)) +
geom_flat_violin(position = position_nudge(x = .1, y = 0), scale = 'width', alpha = 0.2, width = 1.5) +
geom_boxplot(color = 'black', width = .07, outlier.shape = NA, alpha = 0.0, size = 1.0, position = position_nudge(x = .16, y = 0)) +
geom_point(position = position_jitter(width = 0.03, height = 0.02), size = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score",
limits = c(10,30)
) +
scale_x_discrete(
name="Sigma"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette, ) +
scale_fill_manual(values = cb_palette) +
ggtitle('Best performance throughout')+
p_theme
plot_grid(
plot +
theme(legend.position="none"),
legend,
nrow=2,
rel_heights = c(3,1)
)
5.3.2.1 Stats
Summary statistics for the best performance.
performance = filter(best_df, var == 'pop_fit_max' & acro == 'ord')
performance$Sigma = factor(performance$Sigma, levels = c('0','1.2','2.5','0.6','0.3','0.1','5'))
performance %>%
group_by(Sigma) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(val)),
min = min(val / DIMENSIONALITY, na.rm = TRUE),
median = median(val / DIMENSIONALITY, na.rm = TRUE),
mean = mean(val / DIMENSIONALITY, na.rm = TRUE),
max = max(val / DIMENSIONALITY, na.rm = TRUE),
IQR = IQR(val / DIMENSIONALITY, na.rm = TRUE)
)## # A tibble: 7 x 8
## Sigma count na_cnt min median mean max IQR
## <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0 50 0 21.8 23.6 23.6 25.0 0.807
## 2 1.2 50 0 13.8 14.9 15.0 16.5 0.851
## 3 2.5 50 0 13.2 14.7 14.7 16.2 0.727
## 4 0.6 50 0 12.4 14.1 14.1 15.6 0.834
## 5 0.3 50 0 12.9 14.0 13.9 15.6 0.680
## 6 0.1 50 0 12.4 14.1 13.9 15.4 1.04
## 7 5 50 0 10.2 11.6 11.6 13.2 0.973Kruskal–Wallis test illustrates evidence of statistical differences.
##
## Kruskal-Wallis rank sum test
##
## data: val by Sigma
## Kruskal-Wallis chi-squared = 265.47, df = 6, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction.
pairwise.wilcox.test(x = performance$val, g = performance$Sigma, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'l')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: performance$val and performance$Sigma
##
## 0 1.2 2.5 0.6 0.3 0.1
## 1.2 < 2e-16 - - - - -
## 2.5 < 2e-16 0.90 - - - -
## 0.6 < 2e-16 1.3e-07 3.7e-05 - - -
## 0.3 < 2e-16 1.3e-11 4.7e-09 0.95 - -
## 0.1 < 2e-16 2.8e-10 1.2e-07 1.00 1.00 -
## 5 < 2e-16 < 2e-16 < 2e-16 < 2e-16 < 2e-16 3.3e-16
##
## P value adjustment method: bonferroni5.4 Contradictory objectives results
Here we present the results for activation gene coverage and satisfactory trait coverage found by each selection scheme parameter on the contradictory objectives diagnostic. 50 replicates are conducted for each scheme parameters explored.
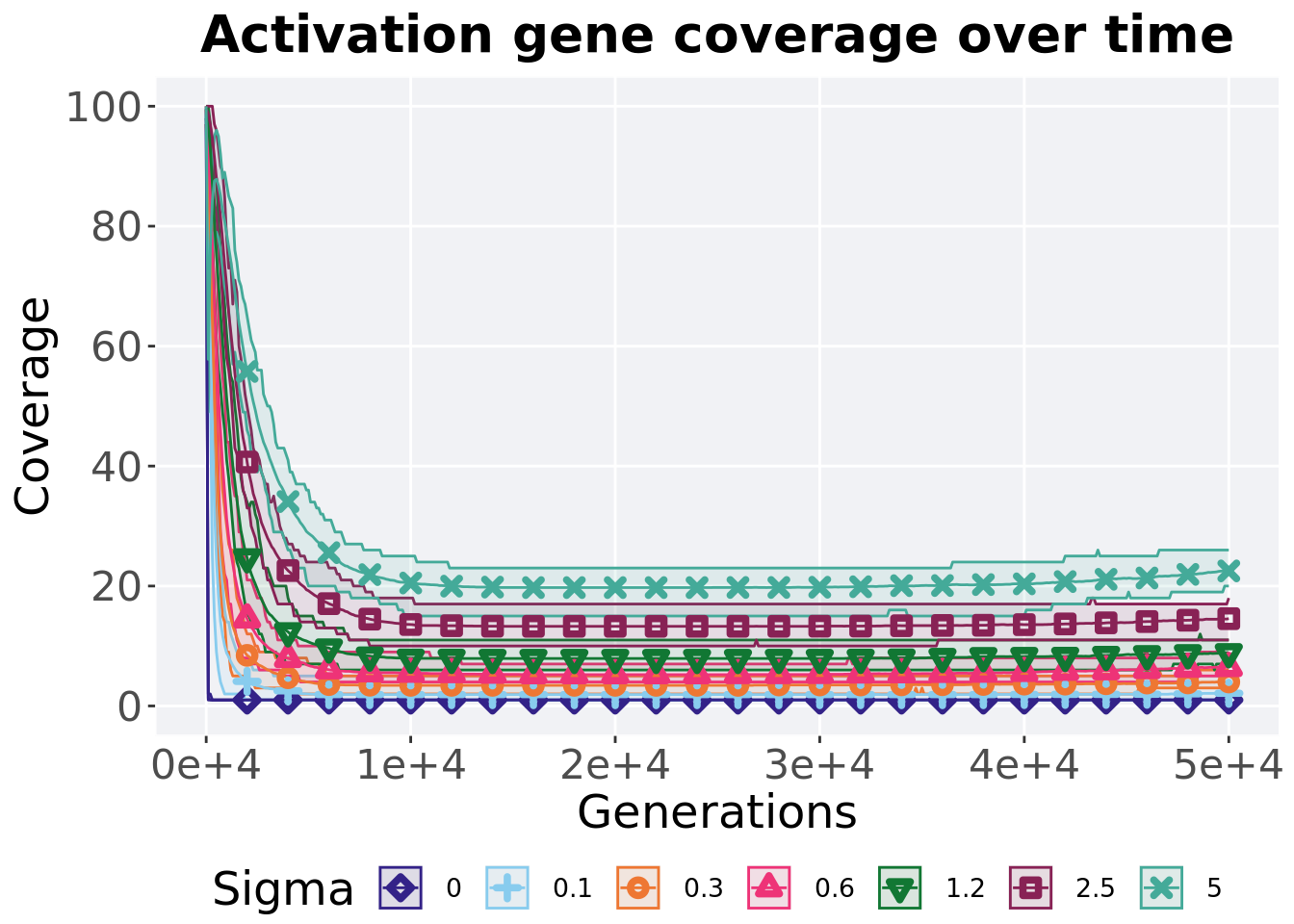
5.4.1 Activation gene coverage over time
Activation gene coverage in a population over time. Data points on the graph is the average activation gene coverage across 50 replicates every 2000 generations. Shading comes from the best and worse coverage across 50 replicates.
lines = filter(over_time_df, acro == 'con') %>%
group_by(Sigma, gen) %>%
dplyr::summarise(
min = min(uni_str_pos),
mean = mean(uni_str_pos),
max = max(uni_str_pos)
)## `summarise()` has grouped output by 'Sigma'. You can override using the
## `.groups` argument.ggplot(lines, aes(x=gen, y=mean, group = Sigma, fill = Sigma, color = Sigma, shape = Sigma)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage",
limits=c(0, 100),
breaks=seq(0,100, 20),
labels=c("0", "20", "40", "60", "80", "100")
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Activation gene coverage over time')+
p_theme +
guides(
shape=guide_legend(nrow=1, title.position = "left", title = 'Sigma'),
color=guide_legend(nrow=1, title.position = "left", title = 'Sigma'),
fill=guide_legend(nrow=1, title.position = "left", title = 'Sigma')
)
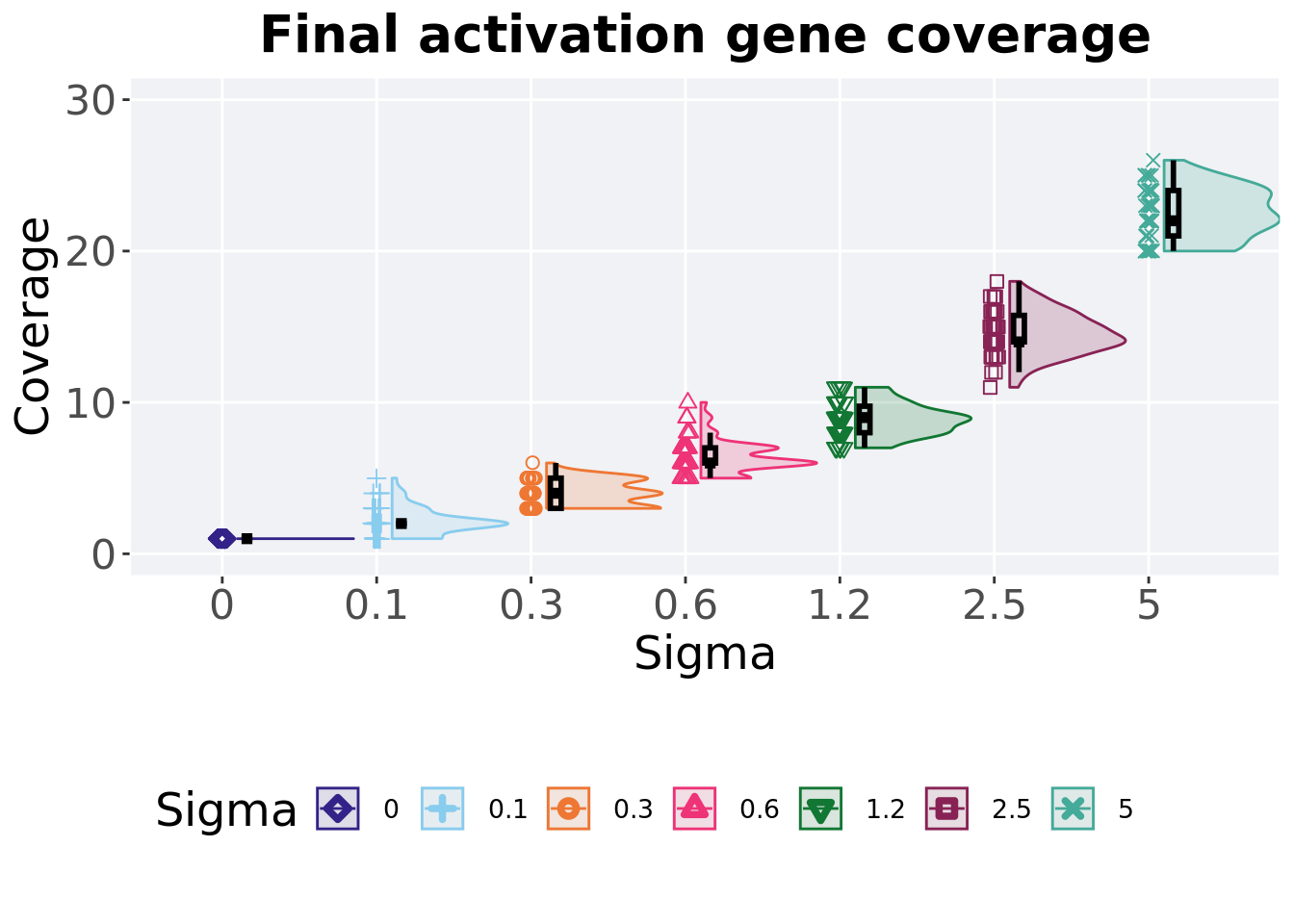
5.4.2 Final activation gene coverage
Activation gene coverage found in the final population at 50,000 generations.
plot = filter(over_time_df, gen == 50000 & acro == 'con') %>%
ggplot(., aes(x = Sigma, y = uni_str_pos, color = Sigma, fill = Sigma, shape = Sigma)) +
geom_flat_violin(position = position_nudge(x = .1, y = 0), scale = 'width', alpha = 0.2, width = 1.5) +
geom_boxplot(color = 'black', width = .07, outlier.shape = NA, alpha = 0.0, size = 1.0, position = position_nudge(x = .16, y = 0)) +
geom_point(position = position_jitter(width = 0.03, height = 0.02), size = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage",
limits=c(0, 30)
) +
scale_x_discrete(
name="Sigma"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette, ) +
scale_fill_manual(values = cb_palette) +
ggtitle('Final activation gene coverage')+
p_theme
plot_grid(
plot +
theme(legend.position="none"),
legend,
nrow=2,
rel_heights = c(3,1)
)
5.4.2.1 Stats
Summary statistics for the generation a satisfactory solution is found.
act_coverage = filter(over_time_df, gen == 50000 & acro == 'con')
act_coverage$Sigma = factor(act_coverage$Sigma, levels = FS_LIST)
act_coverage %>%
group_by(Sigma) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(uni_str_pos)),
min = min(uni_str_pos, na.rm = TRUE),
median = median(uni_str_pos, na.rm = TRUE),
mean = mean(uni_str_pos, na.rm = TRUE),
max = max(uni_str_pos, na.rm = TRUE),
IQR = IQR(uni_str_pos, na.rm = TRUE)
)## # A tibble: 7 x 8
## Sigma count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 0 50 0 1 1 1 1 0
## 2 0.1 50 0 1 2 2.12 5 0
## 3 0.3 50 0 3 4 4 6 2
## 4 0.6 50 0 5 6 6.42 10 1
## 5 1.2 50 0 7 9 8.88 11 1.75
## 6 2.5 50 0 11 14 14.6 18 1.75
## 7 5 50 0 20 22 22.5 26 3Kruskal–Wallis test illustrates evidence of statistical differences.
##
## Kruskal-Wallis rank sum test
##
## data: uni_str_pos by Sigma
## Kruskal-Wallis chi-squared = 337.59, df = 6, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction.
pairwise.wilcox.test(x = act_coverage$uni_str_pos, g = act_coverage$Sigma, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'g')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: act_coverage$uni_str_pos and act_coverage$Sigma
##
## 0 0.1 0.3 0.6 1.2 2.5
## 0.1 1.0e-13 - - - - -
## 0.3 < 2e-16 5.9e-13 - - - -
## 0.6 < 2e-16 < 2e-16 3.3e-15 - - -
## 1.2 < 2e-16 < 2e-16 < 2e-16 3.9e-13 - -
## 2.5 < 2e-16 < 2e-16 < 2e-16 < 2e-16 < 2e-16 -
## 5 < 2e-16 < 2e-16 < 2e-16 < 2e-16 < 2e-16 < 2e-16
##
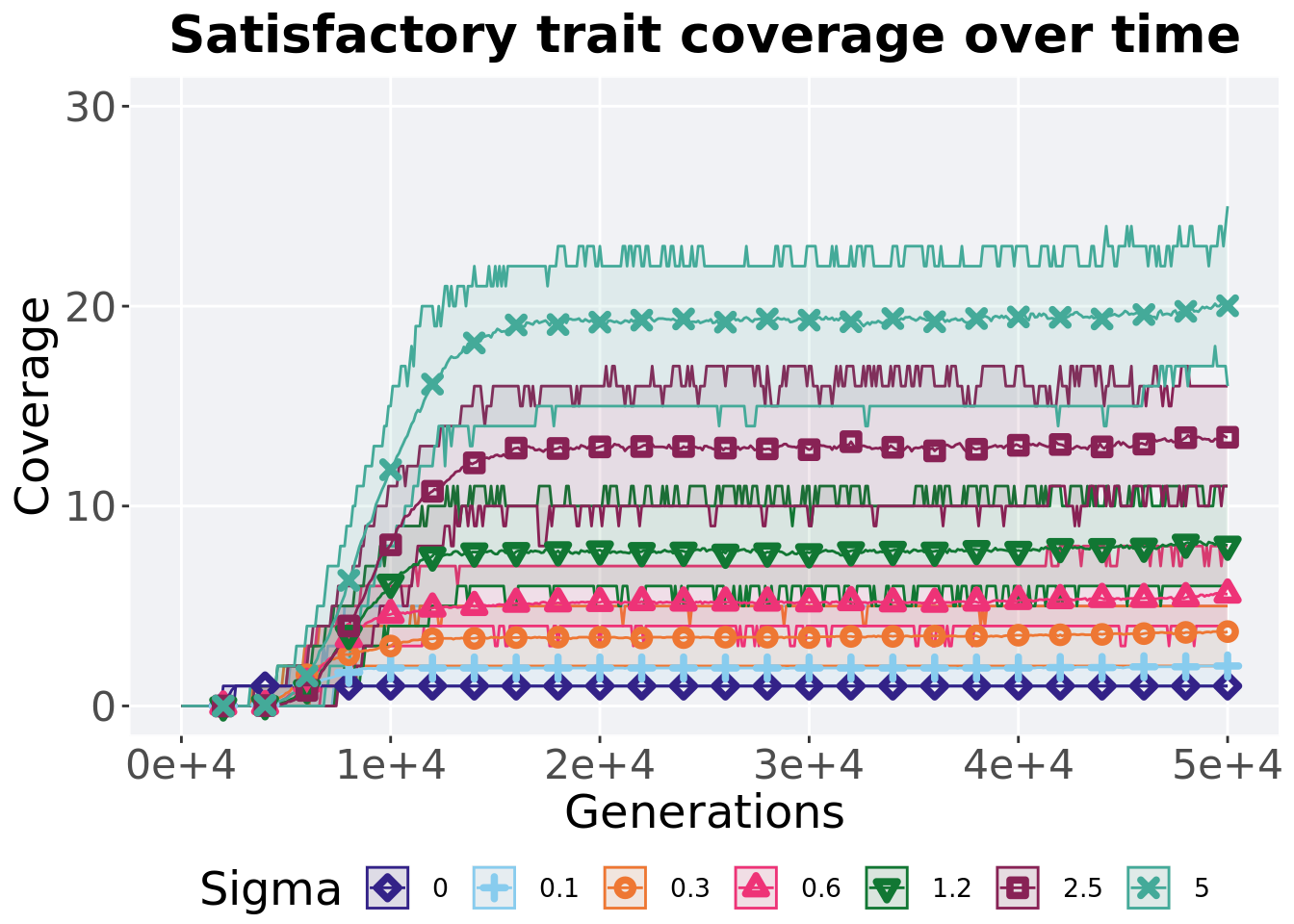
## P value adjustment method: bonferroni5.4.3 Satisfactory trait coverage over time
Satisfactory trait coverage in a population over time. Data points on the graph is the average activation gene coverage across 50 replicates every 2000 generations. Shading comes from the best and worse coverage across 50 replicates.
lines = filter(over_time_df, acro == 'con') %>%
group_by(Sigma, gen) %>%
dplyr::summarise(
min = min(pop_uni_obj),
mean = mean(pop_uni_obj),
max = max(pop_uni_obj)
)## `summarise()` has grouped output by 'Sigma'. You can override using the
## `.groups` argument.ggplot(lines, aes(x=gen, y=mean, group = Sigma, fill = Sigma, color = Sigma, shape = Sigma)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage",
limits=c(0, 30)
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Satisfactory trait coverage over time')+
p_theme +
guides(
shape=guide_legend(nrow=1, title.position = "left", title = 'Sigma'),
color=guide_legend(nrow=1, title.position = "left", title = 'Sigma'),
fill=guide_legend(nrow=1, title.position = "left", title = 'Sigma')
)
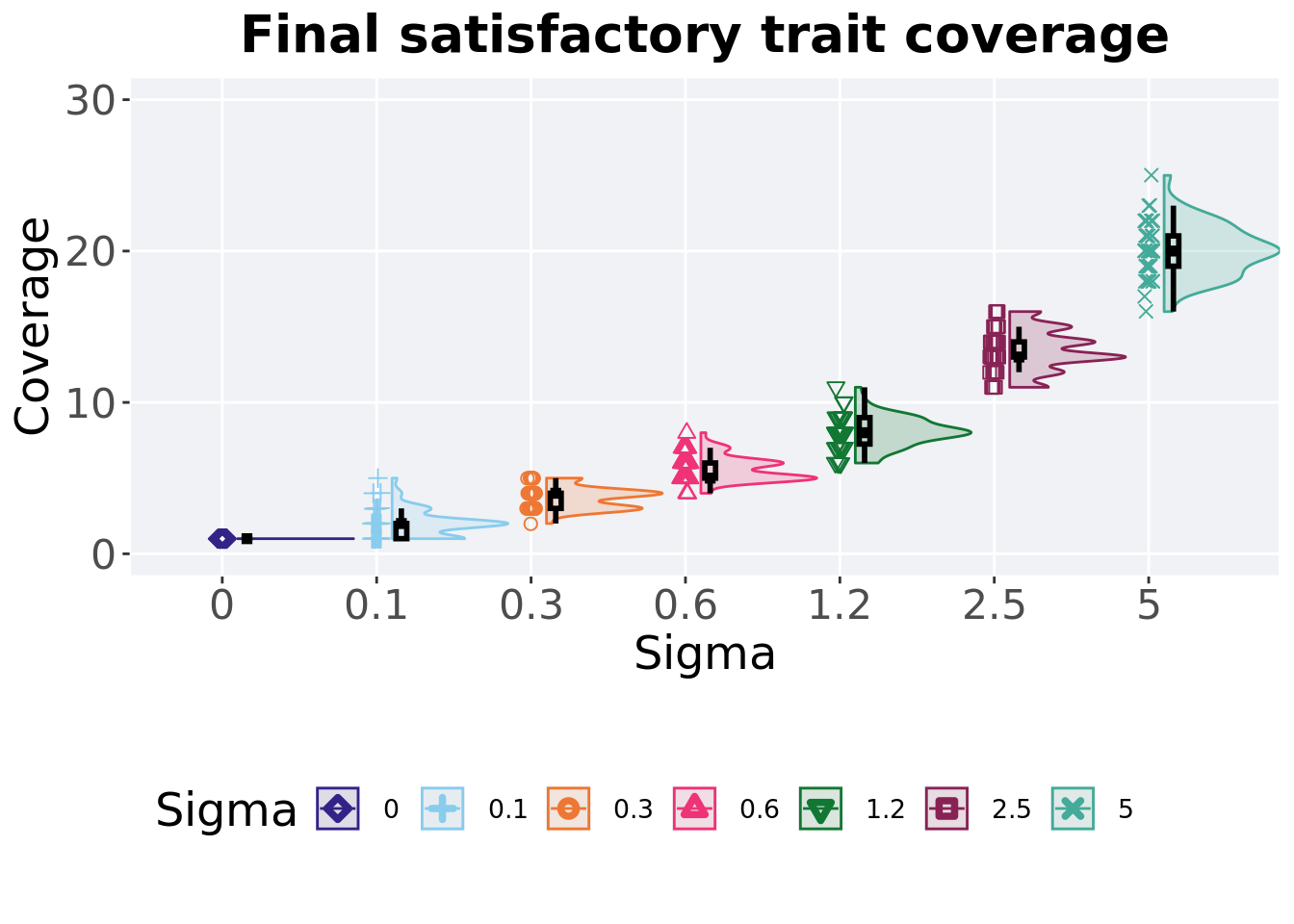
5.4.4 Final satisfactory trait coverage
Satisfactory trait coverage found in the final population at 50,000 generations.
plot = filter(over_time_df, gen == 50000 & acro == 'con') %>%
ggplot(., aes(x = Sigma, y = pop_uni_obj, color = Sigma, fill = Sigma, shape = Sigma)) +
geom_flat_violin(position = position_nudge(x = .1, y = 0), scale = 'width', alpha = 0.2, width = 1.5) +
geom_boxplot(color = 'black', width = .07, outlier.shape = NA, alpha = 0.0, size = 1.0, position = position_nudge(x = .16, y = 0)) +
geom_point(position = position_jitter(width = 0.03, height = 0.02), size = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage",
limits=c(0, 30)
) +
scale_x_discrete(
name="Sigma"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette, ) +
scale_fill_manual(values = cb_palette) +
ggtitle('Final satisfactory trait coverage')+
p_theme
plot_grid(
plot +
theme(legend.position="none"),
legend,
nrow=2,
rel_heights = c(3,1)
)
5.4.4.1 Stats
Summary statistics for the generation a satisfactory solution is found.
sat_coverage = filter(over_time_df, gen == 50000 & acro == 'con')
sat_coverage$Sigma = factor(sat_coverage$Sigma, levels = FS_LIST)
sat_coverage %>%
group_by(Sigma) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(pop_uni_obj)),
min = min(pop_uni_obj, na.rm = TRUE),
median = median(pop_uni_obj, na.rm = TRUE),
mean = mean(pop_uni_obj, na.rm = TRUE),
max = max(pop_uni_obj, na.rm = TRUE),
IQR = IQR(pop_uni_obj, na.rm = TRUE)
)## # A tibble: 7 x 8
## Sigma count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 0 50 0 1 1 1 1 0
## 2 0.1 50 0 1 2 2 5 1
## 3 0.3 50 0 2 4 3.72 5 1
## 4 0.6 50 0 4 5 5.6 8 1
## 5 1.2 50 0 6 8 8.04 11 1.75
## 6 2.5 50 0 11 13 13.4 16 1
## 7 5 50 0 16 20 20.0 25 2Kruskal–Wallis test illustrates evidence of statistical differences.
##
## Kruskal-Wallis rank sum test
##
## data: pop_uni_obj by Sigma
## Kruskal-Wallis chi-squared = 337.1, df = 6, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction.
pairwise.wilcox.test(x = sat_coverage$pop_uni_obj, g = sat_coverage$Sigma, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 'g')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: sat_coverage$pop_uni_obj and sat_coverage$Sigma
##
## 0 0.1 0.3 0.6 1.2 2.5
## 0.1 9.2e-12 - - - - -
## 0.3 < 2e-16 1.4e-12 - - - -
## 0.6 < 2e-16 < 2e-16 1.2e-14 - - -
## 1.2 < 2e-16 < 2e-16 < 2e-16 1.1e-14 - -
## 2.5 < 2e-16 < 2e-16 < 2e-16 < 2e-16 < 2e-16 -
## 5 < 2e-16 < 2e-16 < 2e-16 < 2e-16 < 2e-16 < 2e-16
##
## P value adjustment method: bonferroni5.5 Multi-path exploration results
Here we present the results for best performances and activation gene coverage found by each selection scheme parameter on the multi-path exploration diagnostic. 50 replicates are conducted for each scheme parameter explored.
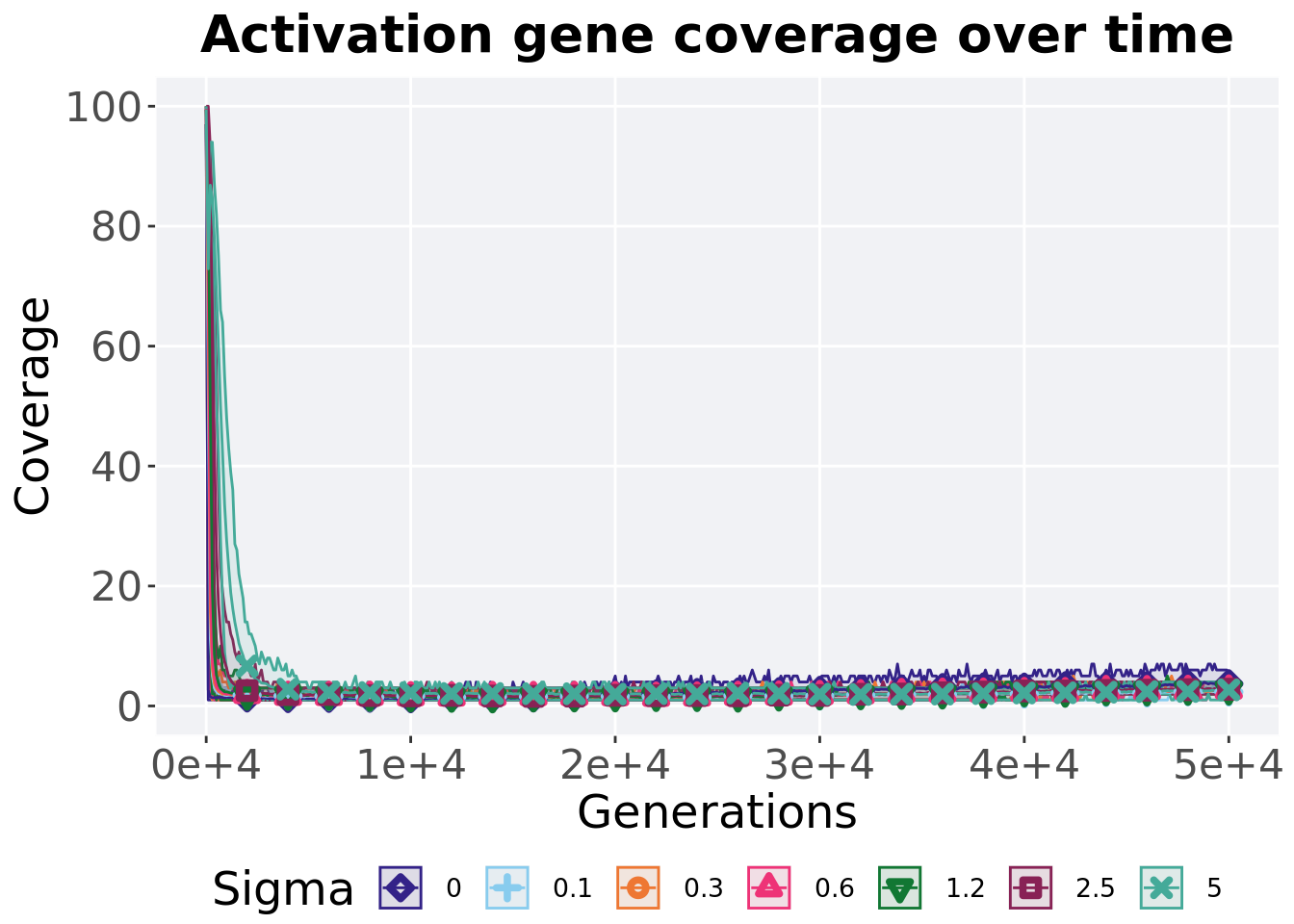
5.5.1 Activation gene coverage over time
Activation gene coverage in a population over time. Data points on the graph is the average activation gene coverage across 50 replicates every 2000 generations. Shading comes from the best and worse coverage across 50 replicates.
lines = filter(over_time_df, acro == 'mpe') %>%
group_by(Sigma, gen) %>%
dplyr::summarise(
min = min(uni_str_pos),
mean = mean(uni_str_pos),
max = max(uni_str_pos)
)## `summarise()` has grouped output by 'Sigma'. You can override using the
## `.groups` argument.ggplot(lines, aes(x=gen, y=mean, group = Sigma, fill = Sigma, color = Sigma, shape = Sigma)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage",
limits=c(0, 100),
breaks=seq(0,100, 20),
labels=c("0", "20", "40", "60", "80", "100")
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Activation gene coverage over time')+
p_theme +
guides(
shape=guide_legend(nrow=1, title.position = "left", title = 'Sigma'),
color=guide_legend(nrow=1, title.position = "left", title = 'Sigma'),
fill=guide_legend(nrow=1, title.position = "left", title = 'Sigma')
)
5.5.2 Final activation gene coverage
Activation gene coverage found in the final population at 50,000 generations.
plot = filter(over_time_df, gen == 50000 & acro == 'mpe') %>%
ggplot(., aes(x = Sigma, y = uni_str_pos, color = Sigma, fill = Sigma, shape = Sigma)) +
geom_flat_violin(position = position_nudge(x = .1, y = 0), scale = 'width', alpha = 0.2, width = 1.5) +
geom_boxplot(color = 'black', width = .07, outlier.shape = NA, alpha = 0.0, size = 1.0, position = position_nudge(x = .16, y = 0)) +
geom_point(position = position_jitter(width = 0.03, height = 0.02), size = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Coverage",
limits=c(0.9, 5.1)
) +
scale_x_discrete(
name="Sigma"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette, ) +
scale_fill_manual(values = cb_palette) +
ggtitle('Final activation gene coverage')+
p_theme
plot_grid(
plot +
theme(legend.position="none"),
legend,
nrow=2,
rel_heights = c(3,1)
)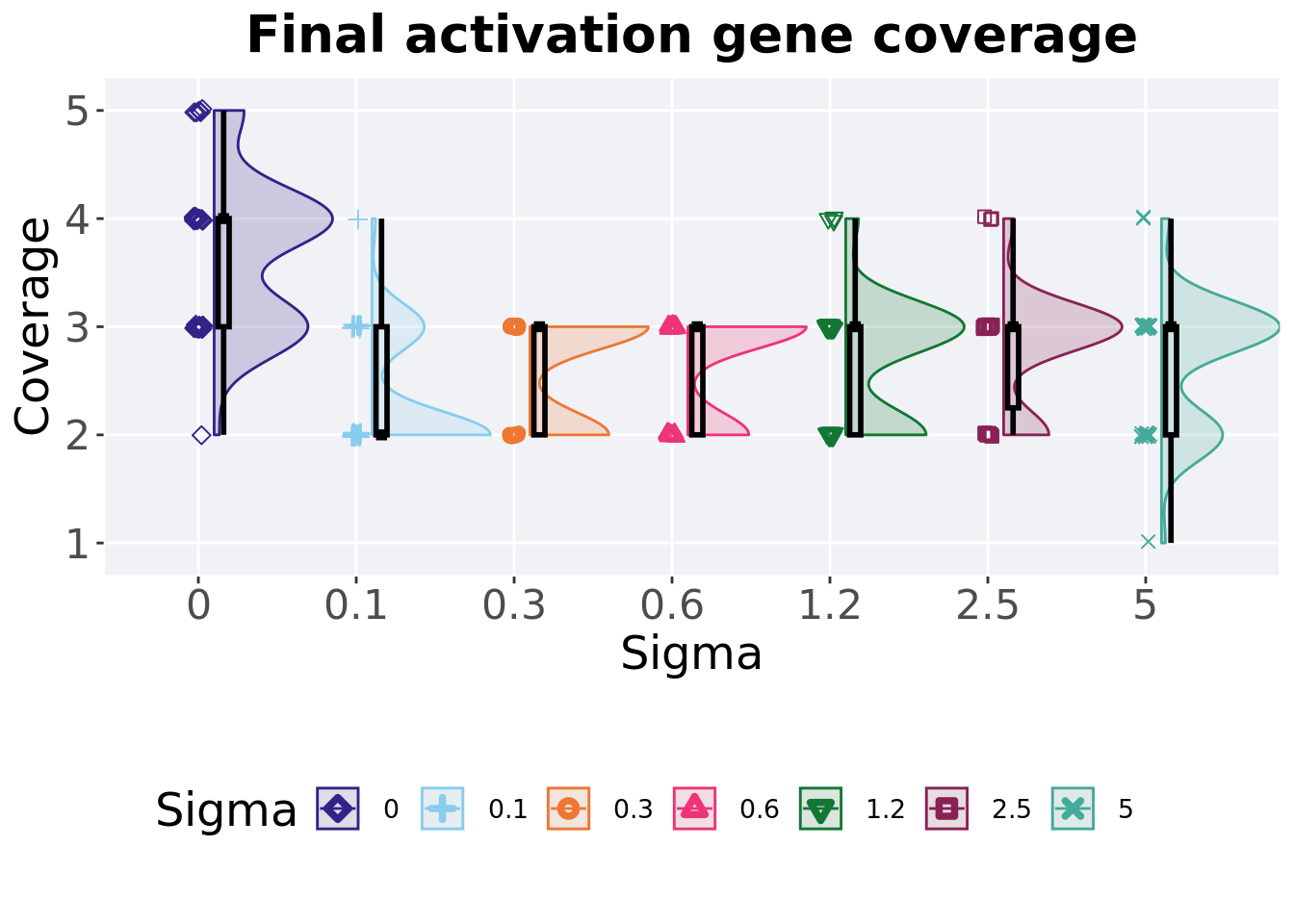
5.5.2.1 Stats
Summary statistics for the generation a satisfactory solution is found.
act_coverage = filter(over_time_df, gen == 50000 & acro == 'mpe')
act_coverage$Sigma = factor(act_coverage$Sigma, levels = FS_LIST)
act_coverage %>%
group_by(Sigma) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(uni_str_pos)),
min = min(uni_str_pos, na.rm = TRUE),
median = median(uni_str_pos, na.rm = TRUE),
mean = mean(uni_str_pos, na.rm = TRUE),
max = max(uni_str_pos, na.rm = TRUE),
IQR = IQR(uni_str_pos, na.rm = TRUE)
)## # A tibble: 7 x 8
## Sigma count na_cnt min median mean max IQR
## <fct> <int> <int> <int> <dbl> <dbl> <int> <dbl>
## 1 0 50 0 2 4 3.7 5 1
## 2 0.1 50 0 2 2 2.34 4 1
## 3 0.3 50 0 2 3 2.6 3 1
## 4 0.6 50 0 2 3 2.66 3 1
## 5 1.2 50 0 2 3 2.68 4 1
## 6 2.5 50 0 2 3 2.8 4 0.75
## 7 5 50 0 1 3 2.68 4 1Kruskal–Wallis test illustrates evidence of statistical differences.
##
## Kruskal-Wallis rank sum test
##
## data: uni_str_pos by Sigma
## Kruskal-Wallis chi-squared = 98.878, df = 6, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction.
pairwise.wilcox.test(x = act_coverage$uni_str_pos, g = act_coverage$Sigma, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 't')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: act_coverage$uni_str_pos and act_coverage$Sigma
##
## 0 0.1 0.3 0.6 1.2 2.5
## 0.1 7.8e-13 - - - - -
## 0.3 1.3e-10 0.16405 - - - -
## 0.6 3.6e-10 0.02571 1.00000 - - -
## 1.2 6.7e-09 0.05563 1.00000 1.00000 - -
## 2.5 4.6e-08 0.00073 1.00000 1.00000 1.00000 -
## 5 4.2e-09 0.03076 1.00000 1.00000 1.00000 1.00000
##
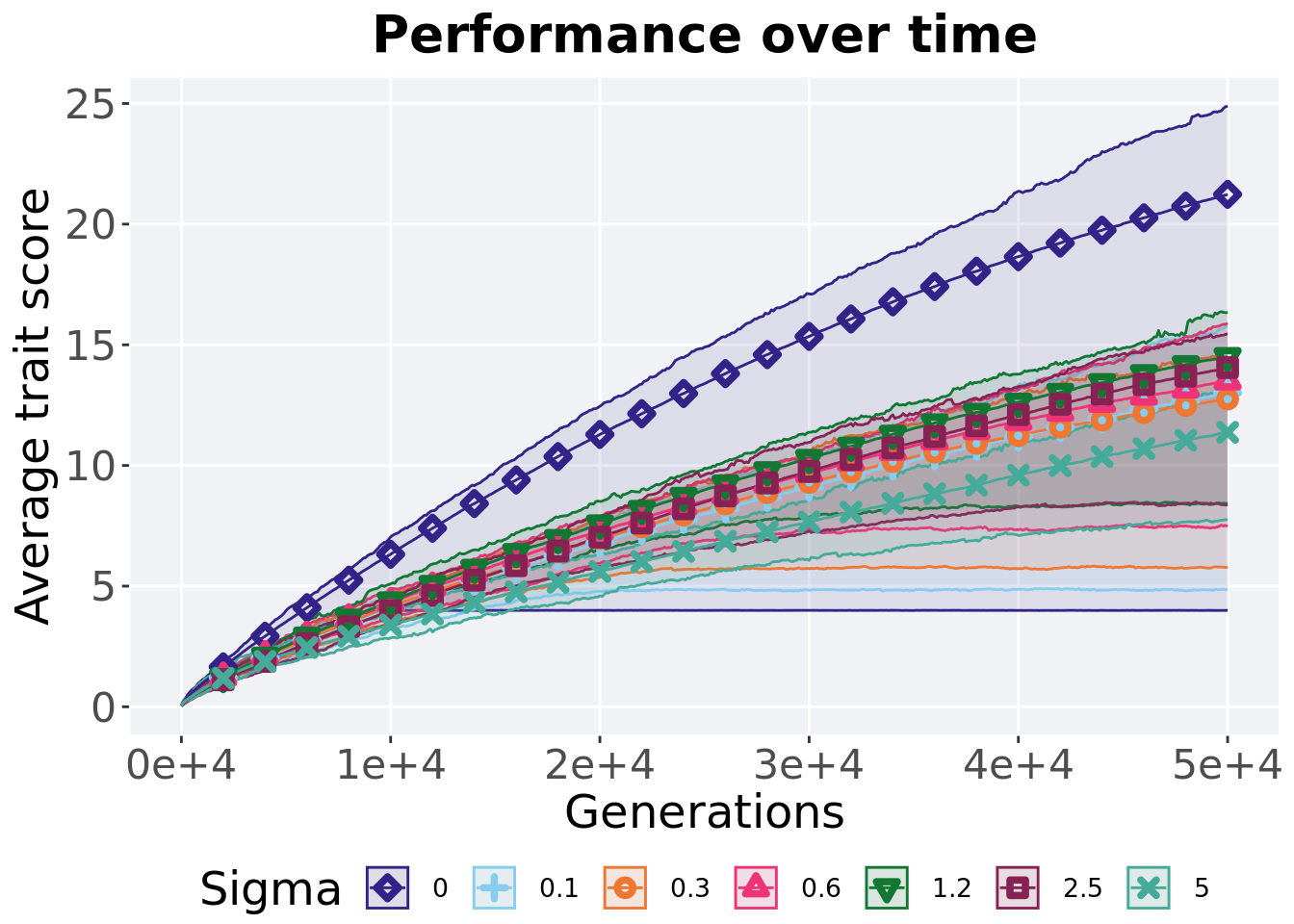
## P value adjustment method: bonferroni5.5.3 Performance over time
Best performance in a population over time. Data points on the graph is the average performance across 50 replicates every 2000 generations. Shading comes from the best and worse performance across 50 replicates.
lines = filter(over_time_df, acro == 'mpe') %>%
group_by(Sigma, gen) %>%
dplyr::summarise(
min = min(pop_fit_max) / DIMENSIONALITY,
mean = mean(pop_fit_max) / DIMENSIONALITY,
max = max(pop_fit_max) / DIMENSIONALITY
)## `summarise()` has grouped output by 'Sigma'. You can override using the
## `.groups` argument.ggplot(lines, aes(x=gen, y=mean, group = Sigma, fill = Sigma, color = Sigma, shape = Sigma)) +
geom_ribbon(aes(ymin = min, ymax = max), alpha = 0.1) +
geom_line(size = 0.5) +
geom_point(data = filter(lines, gen %% 2000 == 0 & gen != 0), size = 1.5, stroke = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score"
) +
scale_x_continuous(
name="Generations",
limits=c(0, 50000),
breaks=c(0, 10000, 20000, 30000, 40000, 50000),
labels=c("0e+4", "1e+4", "2e+4", "3e+4", "4e+4", "5e+4")
) +
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette) +
scale_fill_manual(values = cb_palette) +
ggtitle('Performance over time')+
p_theme +
guides(
shape=guide_legend(nrow=1, title.position = "left", title = 'Sigma'),
color=guide_legend(nrow=1, title.position = "left", title = 'Sigma'),
fill=guide_legend(nrow=1, title.position = "left", title = 'Sigma')
)
5.5.4 Best performance throughout
Best performance reached throughout 50,000 generations in a population.
plot = filter(best_df, var == 'pop_fit_max' & acro == 'mpe') %>%
ggplot(., aes(x = Sigma, y = val / DIMENSIONALITY, color = Sigma, fill = Sigma, shape = Sigma)) +
geom_flat_violin(position = position_nudge(x = .1, y = 0), scale = 'width', alpha = 0.2, width = 1.5) +
geom_boxplot(color = 'black', width = .07, outlier.shape = NA, alpha = 0.0, size = 1.0, position = position_nudge(x = .16, y = 0)) +
geom_point(position = position_jitter(width = 0.03, height = 0.02), size = 2.0, alpha = 1.0) +
scale_y_continuous(
name="Average trait score",
limits=c(0, 25)
) +
scale_x_discrete(
name="Sigma"
)+
scale_shape_manual(values=SHAPE)+
scale_colour_manual(values = cb_palette, ) +
scale_fill_manual(values = cb_palette) +
ggtitle('Best performance throughout')+
p_theme
plot_grid(
plot +
theme(legend.position="none"),
legend,
nrow=2,
rel_heights = c(3,1)
)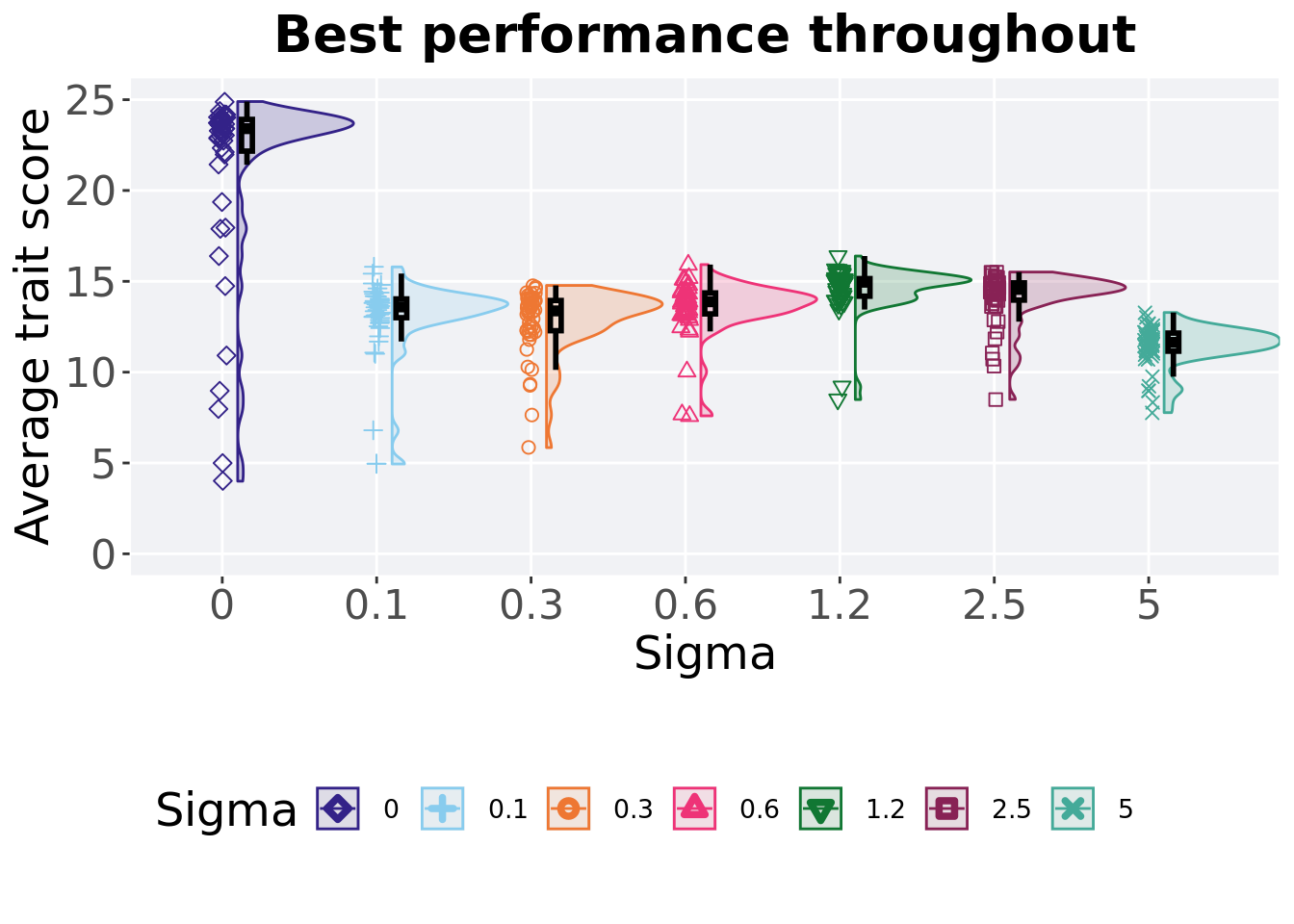
5.5.4.1 Stats
Summary statistics for the best performance.
performance = filter(best_df, var == 'pop_fit_max' & acro == 'mpe')
performance$Sigma = factor(performance$Sigma, levels = c('0','2.5','1.2','0.6','0.3','0.1','5'))
performance %>%
group_by(Sigma) %>%
dplyr::summarise(
count = n(),
na_cnt = sum(is.na(val)),
min = min(val / DIMENSIONALITY, na.rm = TRUE),
median = median(val / DIMENSIONALITY, na.rm = TRUE),
mean = mean(val / DIMENSIONALITY, na.rm = TRUE),
max = max(val / DIMENSIONALITY, na.rm = TRUE),
IQR = IQR(val / DIMENSIONALITY, na.rm = TRUE)
)## # A tibble: 7 x 8
## Sigma count na_cnt min median mean max IQR
## <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0 50 0 4.00 23.4 21.3 24.9 1.74
## 2 2.5 50 0 8.50 14.6 14.1 15.5 0.949
## 3 1.2 50 0 8.49 15.0 14.6 16.4 0.967
## 4 0.6 50 0 7.60 13.9 13.6 15.9 1.15
## 5 0.3 50 0 5.85 13.4 12.8 14.8 1.66
## 6 0.1 50 0 4.95 13.6 13.1 15.8 1.03
## 7 5 50 0 7.77 11.6 11.4 13.3 0.997Kruskal–Wallis test illustrates evidence of statistical differences.
##
## Kruskal-Wallis rank sum test
##
## data: val by Sigma
## Kruskal-Wallis chi-squared = 183.09, df = 6, p-value < 2.2e-16Results for post-hoc Wilcoxon rank-sum test with a Bonferroni correction.
pairwise.wilcox.test(x = performance$val, g = performance$Sigma, p.adjust.method = "bonferroni",
paired = FALSE, conf.int = FALSE, alternative = 't')##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: performance$val and performance$Sigma
##
## 0 2.5 1.2 0.6 0.3 0.1
## 2.5 2.3e-10 - - - - -
## 1.2 4.5e-10 0.59041 - - - -
## 0.6 1.1e-10 0.02305 1.0e-05 - - -
## 0.3 7.5e-11 3.9e-06 5.4e-10 0.15225 - -
## 0.1 8.3e-11 0.00026 3.5e-08 1.00000 1.00000 -
## 5 6.5e-11 1.3e-11 3.4e-14 2.8e-12 2.3e-07 1.0e-09
##
## P value adjustment method: bonferroni